Uncovering antibiotic resistance: extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa from dipteran flies in residential dumping and livestock environments
- PMID: 40535008
- PMCID: PMC12174077
- DOI: 10.3389/fmicb.2025.1586811
Uncovering antibiotic resistance: extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa from dipteran flies in residential dumping and livestock environments
Abstract
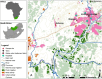
Introduction: Pseudomonas aeruginosa poses challenges in clinical and environmental contexts due to its capacity to colonize natural ecosystems and antibiotic resistance. This study characterized P. aeruginosa harboured by Diptera flies collected from illegal residential dumping sites and livestock (cattle, sheep, and goats) kraals in Potchefstroom, South Africa.
Methods: The P. aeruginosa isolates were characterized using classical microbiological tests and species-specific gyrase B gene PCR assay. Antibiotic resistance (AR) was assessed on the isolates using disc diffusion assay (DDA). Additionally, PCR screened six virulence genes (exoS, plcN, plcH, toxA, lasB, and algD) among the isolates. Whole genome sequencing (WGS) was employed to confirm the identity and determine antibiotic resistance genes (ARGs) on selected isolates.
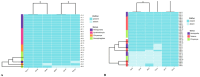
Results: Culture-based and molecular assays showed that P. aeruginosa is prevalent in Diptera flies (Hemipyrellia spp., Synthesiomya spp., Chrysomya spp., Sarchophagidae spp., and Tabanus spp.) from livestock kraals (75%; n = 36/48) and dumping sites (48%; n = 23/48). The most detected virulent gene among the isolates was exoS (96.6%), followed by plcN and algD genes (83.1%), lasB (81.4%), toxA (76.3%), and plcH (47.5%). All P. aeruginosa isolates were resistant to metronidazole, sulphamethoxazole, cefazolin and amoxicillin based on DDA. The sulfonamide resistance sulI gene (88.1%) was the most detected ARG from the P. aeruginosa isolates, followed by acc(3)-IV (80.6%) coding for aminoglycoside. WGS revealed that P. aeruginosa isolates belong to the sequence type (ST3808), which is multidrug-resistant and contains ARGs for fosfomycin (fosA), ampicillin (bla OXA-50), chloramphenicol (catB7), beta-lactamase (bla PAO), and aminoglycoside (aph(3')-IIb).
Discussion: This study isolated ESBL-producing P. aeruginosa from various Diptera fly species collected from livestock kraals and residential dumping sites. This bacterium is important to "One Health" due to its multidrug resistance character and zoonotic nature. As a result, it requires consolidated control and management policies from the environmental, veterinary, and human health sectors.
Keywords: Diptera flies; Pseudomonas aeruginosa; WGS; antibiotic resistance; virulence genes.
Copyright © 2025 de Wet, Matle, Thekisoe, Lekota and Ramatla.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abdalhamed A., Zeedan G. G., Hussein H., Abdeen E. (2016). Molecular and biochemical characterization of Pseudomonas species isolated from subclinical mastitis milk and ice cream and its susceptibility to allium sativum and commiphorra molmol plant extracts in Egypt. Int. J. Adv. Res. 4, 1669–1679. doi: 10.21474/IJAR01/1378 - DOI
-
- Andrews S. (2017). FastQC: A quality control tool for high throughput sequence data, vol. 2010. Available at: https://cir.nii.ac.jp/crid/1370584340724053142
-
- Ahmadi N., Salimizand H., Zomorodi A. R., Abbas J. E., Ramazanzadeh R., Haghi F., et al. (2024). Genomic diversity of β-lactamase producing Pseudomonas aeruginosa in Iran; the impact of global high-risk clones. Ann. Clin. Microbiol. Antimicrob. 23:5. doi: 10.1186/s12941-024-00668-5 - DOI - PMC - PubMed
-
- Badawy B., Moustafa S., Shata R., Sayed-Ahmed M. Z., Alqahtani S. S., Ali M. S., et al. (2023). Prevalence of multidrug-resistant Pseudomonas aeruginosa isolated from dairy cattle, Milk, environment, and workers’ hands. Microorganisms 11:2775. doi: 10.3390/microorganisms11112775, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

