Comprehensive assessment of AlphaFold's predictions of secondary structure and solvent accessibility at the amino acid-level in eukaryotic, bacterial and archaeal proteins
- PMID: 40535106
- PMCID: PMC12173809
- DOI: 10.1016/j.csbj.2025.05.047
Comprehensive assessment of AlphaFold's predictions of secondary structure and solvent accessibility at the amino acid-level in eukaryotic, bacterial and archaeal proteins
Abstract
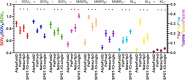
Numerous sequence-based predictors of the amino acid (AA)-level solvent accessibility (SA) and secondary structure (SS) of proteins have been developed. We empirically investigated whether these two key characteristics of AA-level structure can be accurately predicted from putative structures generated by the popular AlphaFold2. We compared AlphaFold2's results against several representative SS and SA predictors on a large test dataset that covers five distinct taxonomic groups (animals, plants, fungi, bacteria, and archaea). We used a broad collection of metrics that evaluate predictions of the numeric and binary (buried vs. solvent exposed) SA and the 3-state SS at both AA- and SS-region levels. We found that AlphaFold2 generated very accurate results, with high average Q3 accuracy of 0.928 for the SS prediction and high Pearson Correlation Coefficient (PCC) of 0.815 between its putative and native SA values. AlphaFold2 significantly and consistently outperforms the considered predictors of SA and SS across the five taxonomic groups and both AA and region level evaluations. Moreover, we demonstrated that AlphaFold2 nearly perfectly reconstructs distributions of the sizes and numbers of the SS regions. We also showed that AlphaFold2 substantially improves over the SS and SA predictors when tested on a low sequence similarity test dataset, although its results and results of two other predictors suffer a modest drop in the quality of predicting SS regions. Altogether, our results suggest that AlphaFold2 makes very accurate predictions of SS and SA, which can be easily extracted from 200+ million pre-computed AF2's structure predictions in AlphaFoldDB.
Keywords: AlphaFold; Evaluation; Prediction; Protein structure; Secondary structure; Solvent accessibility.
© 2025 The Authors.
Conflict of interest statement
Authors declare no conflict of interests.
Figures


Similar articles
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
AlphaFold2's training set powers its predictions of some fold-switched conformations.Protein Sci. 2025 Apr;34(4):e70105. doi: 10.1002/pro.70105. Protein Sci. 2025. PMID: 40130805 Free PMC article.
-
Prediction, screening and characterization of novel bioactive tetrapeptide matrikines for skin rejuvenation.Br J Dermatol. 2024 Jun 20;191(1):92-106. doi: 10.1093/bjd/ljae061. Br J Dermatol. 2024. PMID: 38375775
-
Chemoautotrophy in subzero environments and the potential for cold-adapted Rubisco.Appl Environ Microbiol. 2025 Jun 18;91(6):e0060425. doi: 10.1128/aem.00604-25. Epub 2025 May 30. Appl Environ Microbiol. 2025. PMID: 40444981 Free PMC article.
-
Prenatal administration of progestogens for preventing spontaneous preterm birth in women with a multiple pregnancy.Cochrane Database Syst Rev. 2019 Nov 20;2019(11):CD012024. doi: 10.1002/14651858.CD012024.pub3. Cochrane Database Syst Rev. 2019. PMID: 31745984 Free PMC article.
References
LinkOut - more resources
Full Text Sources
