Encoding Genes of Metallo β-Lactamases (IMP, NDM, and VIM) in Klebsiella Pneumoniae in Iran: A Systematic Review and Meta-Analysis
- PMID: 40535515
- PMCID: PMC12174657
- DOI: 10.1002/hsr2.70927
Encoding Genes of Metallo β-Lactamases (IMP, NDM, and VIM) in Klebsiella Pneumoniae in Iran: A Systematic Review and Meta-Analysis
Abstract
Background and aim: Antimicrobial resistance is one of the top 10 global public health threats, particularly growing resistance to β-lactam antibiotics, including carbapenems, which are among the last-resort antibiotics used against multidrug and extensively drug-resistant bacteria.
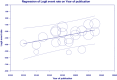
Methods: This meta-analysis explores the status of class B carbapenemases, including Imipenemase (IMP), New Delhi metallo-β-lactamase (NDM), and Verona integron-encoded metallo-β-lactamase (VIM) in Klebsiella pneumoniae in Iran. Eligible articles were retrieved from electronic literature searches of online databases (PubMed, Scopus, Wiley Online Library, ScienceDirect, and Google Scholar) to identify studies published between 2010 and 2024.
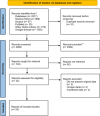
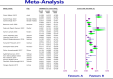
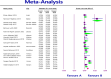
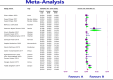
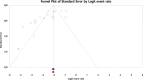
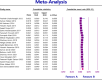
Results: The study selection process yielded 2517 studies. A total of 124 studies fulfilled the inclusion criteria; of these, 25 studies were eligible for meta-analysis. A meta-analysis of prevalence studies demonstrated that the most encoding genes of MBLs were to NDM, VIM, and IMP at 7.1% (95% confidence interval [CI]: 4.9%-10.3%), 1.9% (95% CI: 0.9%-4.1%), and 0.9% (95% CI: 0.4%-2.0%), with heterogeneity (I-squared [I 2] = 86.565, I 2 = 88.460, I 2 = 73.835, p < 0.0001 for all). The evaluated pooled prevalence of NDM, VIM, and IMP was highest in Arak (29.8%; 95% CI: 22.3%-38.5%), Babol (30.0%; 95% CI: 19.0%-44.0%), and Dezful (18.8%; 95% CI: 8.7%-35.9%), respectively.
Conclusion: The increasing prevalence of NDM among encoding genes of MBL in Klebsiella pneumoniae in Iran represents a major clinical obstacle to therapeutic options in the future. These findings highlight the urgent need for effective infection control measures and the development of new treatment strategies to address the rising resistance. Enhanced surveillance and the implementation of targeted antibiotic stewardship programs are crucial in controlling the spread of these resistant strains. However, the study has limitations, including the potential biases related to publication bias and heterogeneity across studies, which should be considered when interpreting the findings.
Keywords: Klebsiella pneumonia; metallo‐β‐lactamases; microbial drug resistance.
© 2025 The Author(s). Health Science Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Organization W. H., WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance (World Health Organization, 2024).
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

