From transmission to adaptive evolution: genomic surveillance of Getah virus
- PMID: 40535537
- PMCID: PMC12174390
- DOI: 10.3389/fcimb.2025.1513392
From transmission to adaptive evolution: genomic surveillance of Getah virus
Abstract
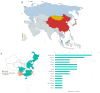
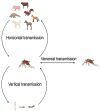
Getah virus (GETV) is a member of the Alphavirus of the Togaviridae. It is a single-stranded positive-RNA virus that is mainly transmitted by mosquitoes. In recent years, the spread of GETV has become increasingly serious, causing serious losses to the animal economy and posing a potential threat to public health. GETV infected animals extend from traditional domestic animals such as horses and pigs to cattle, foxes and other animals. Especially in China, the virus has been detected in many provinces in recent years. In addition, GETV-specific antibodies were detected in healthy humans. However, the threat posed by GETV in China has not received enough attention. In this study, we downloaded all available GETV genome-wide serials (82 serials in total) from the NCBI as of December 2023. We integrate multiple bioinformatics approaches to understand the characteristics of GETV from the perspectives of epidemiology, virus-host co-evolution, and viral adaptation analysis. The results of this study show that GETV is rapidly expanding its host range and geographical distribution at high evolutionary rates due to the lack of commercially available vaccines. Second, we clearly reveal the cross-species transmission of GETV. Finally, we identified important adaptive and active selection sites. GETV and its media are widely distributed in China, and new host infections continue to appear. Therefore, strengthening surveillance and prevention to avoid serious losses to the pandemic is an important task we face today.
Keywords: Getah virus; adaptive evolution; cross-species; epidemiological situation; host range.
Copyright © 2025 Yuan, Hao, Peng, Zhang, Ma, Xiao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Outbreak and epidemic of Getah virus infection in swine by virulence-enhanced GIII variant in Henan, central China in 2024.Virulence. 2025 Dec;16(1):2530661. doi: 10.1080/21505594.2025.2530661. Epub 2025 Jul 13. Virulence. 2025. PMID: 40653751 Free PMC article.
-
Establishment and application of an indirect ELISA for Getah virus E2 antibody detection.J Virol Methods. 2024 Apr;325:114885. doi: 10.1016/j.jviromet.2024.114885. Epub 2024 Jan 14. J Virol Methods. 2024. PMID: 38228247
-
Establishment of reverse transcription recombinase-aided amplification with lateral flow dipstick for the rapid visual detection of Getah virus.Front Cell Infect Microbiol. 2025 Aug 13;15:1631048. doi: 10.3389/fcimb.2025.1631048. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40880631 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Mayaro Virus: An Emerging Alphavirus in the Americas.Viruses. 2024 Aug 14;16(8):1297. doi: 10.3390/v16081297. Viruses. 2024. PMID: 39205271 Free PMC article. Review.
References
-
- Azerigyik F. A., Faizah A. N., Kobayashi D., Amoa-Bosompem M., Matsumura R., Kai I., et al. (2023). Evaluating the mosquito host range of Getah virus and the vector competence of selected medically important mosquitoes in Getah virus transmission. Parasit Vectors. 16, 99. doi: 10.1186/s13071-023-05713-4 - DOI - PMC - PubMed
-
- Bannai H., Nemoto M., Niwa H., Murakami S., Tsujimura K., Yamanaka T., et al. (2017). Geospatial and temporal associations of Getah virus circulation among pigs and horses around the perimeter of outbreaks in Japanese racehorses in 2014 and 2015. BMC Vet. Res. 13, 187. doi: 10.1186/s12917-017-1112-6 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

