Exploring the evolutionary journey of the lumpy skin disease virus through the phylogenetic and phylo-geo network analysis
- PMID: 40535539
- PMCID: PMC12174415
- DOI: 10.3389/fcimb.2025.1575538
Exploring the evolutionary journey of the lumpy skin disease virus through the phylogenetic and phylo-geo network analysis
Abstract
Introduction: Lumpy Skin Disease Virus (LSDV), an emerging pathogen from the Capripoxvirus genus, continues to challenge global livestock health with its expanding host range and genetic adaptability.
Materials and methods: In this study, we report the first isolation and whole genome sequencing of LSDV from Bos frontalis, a semi-domesticated bovine species native to Northeast India, along with the assembly of an isolate from cattle.
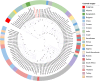
Results: Time to the Most Recent Common Ancestor (TMRCA) estimates support a relatively recent common origin for Indian strains, pointing to ongoing virus circulation and regional adaptation. The maximum likelihood phylogenetic tree of the whole genome and G protein-coupled chemokine receptor (GPCR) gene further demonstrated the clustering of global strains, emphasizing the virus's transboundary movement and genomic diversity. To strengthen phylogenetic inference, we identified shared SNPs, synonymous and non-synonymous mutations across the genome with a total of 2212 variants. Haplotype network and mutation pattern analyses across global genomes further highlighted the conservative evolution of Indian isolates within a distinct haplogroup.
Discussion: Several mutation events between haplogroups highlight the virus's continuous genetic diversification, which correlates with known patterns of spread.
Keywords: TMRCA; genetic diversity; haplotype network; lumpy skin disease virus; phylogenetic analysis; transboundary spread.
Copyright © 2025 Bayyappa, Pyatla, Pabbineedi, Gunturu, Peela, Nagaraj, Tadakod, Gandham and Gulati.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor RMA declared a shared parent affiliation with the author(s) MB, SPa, SN, ST, and BG at the time of review.
Figures




References
-
- Al-Salihi K. (2014). Lumpy skin disease: Review of literature. Mirror Res. Vet. Sci. anim. 3, 6–23.
-
- Amin D. M., Shehab G., Emran R., Hassanien R. T., Alagmy G. N., Hagag N. M., et al. (2021). Diagnosis of naturally occurring lumpy skin disease virus infection in cattle using virological, molecular, and immunohistopathological assays. Vet. World 14, 2230. doi: 10.14202/vetworld.2021.2230-2237 - DOI - PMC - PubMed
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed September 19, 2024).
MeSH terms
LinkOut - more resources
Full Text Sources

