Molecular and epidemiological characterization of carbapenem-resistant hypervirulent Klebsiella pneumoniae in Huaian, China (2022-2024): a retrospective study
- PMID: 40535540
- PMCID: PMC12174136
- DOI: 10.3389/fcimb.2025.1569004
Molecular and epidemiological characterization of carbapenem-resistant hypervirulent Klebsiella pneumoniae in Huaian, China (2022-2024): a retrospective study
Abstract
Objectives: Carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) poses a significant public health challenge. This study investigated the molecular epidemiology, antimicrobial resistance patterns, clinical characteristics, and risk factors of CR-hvKP infection in Huaian, China.
Methods: We retrospectively studied patients infected with carbapenem-resistant K. pneumoniae (CRKP) between November 2022 and September 2024. Whole-genome sequencing was used to detect carbapenemase, virulence, capsular serotype-related genes, and plasmid types in 374 CRKP isolates.
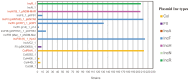
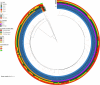
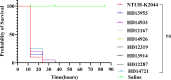
Results: Among them, 57.49% (215/374) strains met the criteria for CR-hvKP. The most common type was blaKPC-2-producing ST11(98.60%, 212/215), whereas K64 (56.74%, 122/215) and KL25 (39.53%, 85/215) were the main capsular serotypes. The CR-hvKP strains showed significantly higher resistance to the tested antibiotics, except for ceftazidime/avibactam and colistin. Resistance rates of CR-hvKP to the three tested antibiotics (minocycline, cotrimoxazole, and amikacin) were higher than those of CRnon-hvKP. Phylogenetic analysis based on whole-genome single-nucleotide polymorphisms divided the 251 isolates into four independent branches, with branch 2 being the most prevalent, indicating high clonality among the strains. Multivariate analysis showed diabetes [odds ratio (OR) = 3.771] and surgery (OR =2.042) to be independent variables associated with CR-hvKP infection.
Conclusions: Notably, the ST11 lineage carrying blaKPC-2 has emerged as a dominant high-risk clone in Huaian. Given the wide distribution of these novel CR-hvKP isolates, global monitoring and stricter control measures should be implemented to prevent their further spread in hospital settings.
Keywords: carbapenem-resistant hypervirulent Klebsiella pneumoniae; epidemiological characterization; molecular characterization; multidrug resistance; phylogenetic analysis; risk factors.
Copyright © 2025 Lian, Li, Peng, Lin, Du, Tang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- CLSI (2021). Performance standards for antimicrobial susceptibility testing; 31th informational supplement (CLSI M100-S31) (Wayne: Clinical and Laboratory Standards Institute; ).
-
- Cui X., Shan B., Zhang X., Qu F., Jia W., Huang B., et al. (2020). Reduced ceftazidime-avibactam susceptibility in KPC-producing klebsiella pneumoniae from patients without ceftazidime-avibactam use history - A multicenter study in China. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01365 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

