Salivary microbiota and IgA responses are different in pre-diabetic individuals compared to normoglycemic controls
- PMID: 40535541
- PMCID: PMC12174153
- DOI: 10.3389/fcimb.2025.1591285
Salivary microbiota and IgA responses are different in pre-diabetic individuals compared to normoglycemic controls
Abstract
Introduction: In recent years, changes in the oral microbiota of patients with type 2 diabetes mellitus (T2DM) have been increasingly recognized. The salivary microbiota may also be altered in pre-diabetes, which is the earliest stage of abnormal blood glucose regulation and a reversible stage preceding T2DM; however, its characteristics are poorly understood. Salivary immunoglobulin A (IgA) is a host defense factor central to the oral immune system and may play an important role in regulating the salivary microbiota. Given that alterations in immunoreactivity are observed in pre-diabetes, we hypothesized that the salivary IgA response may also be altered; however, limited knowledge exists regarding this. Therefore, in the present study, we aimed to evaluate the characteristics of salivary microbiota and IgA responses against salivary microbiota in individuals with pre-diabetes, comparing them to those in individuals with normoglycemia.
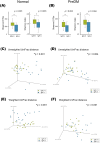
Methods: Saliva samples were collected from 101 pre-diabetic individuals (PreDM group) and 101 age- and sex-matched normoglycemic controls (Normal group). Further, 16S rRNA metagenomic analysis was performed to compare bacterial microbiota composition. For each of the 19 saliva samples from the PreDM and Normal groups, IgA-enriched and IgA-nonenriched fractions were separated via magnetic-activated cell sorting, followed by 16S rRNA metagenomic analysis. The IgA index was calculated to evaluate the difference in the IgA response to each bacterium between the PreDM and Normal groups.
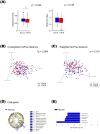
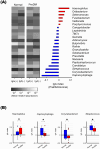
Results: Bacterial species richness was significantly lower in the PreDM group than in the Normal group (observed operational taxonomic unit index, p = 0.042), and a difference between these groups was noted in the overall salivary microbiota structure (unweighted UniFrac distances, p = 0.009). Salivary IgA responses against several bacterial genera differed between the PreDM and Normal groups. Significantly higher IgA responses were noted against Haemophilus in the PreDM group, with lower responses against Capnocytophaga, Corynebacterium, and Streptococcus relative to those in the Normal group.
Conclusions: Salivary microbiota and IgA responses differ between pre-diabetic individuals and normoglycemic controls. The current findings advance our understanding of the interaction between oral bacteria and host immune responses in patients with a poor glycemic status.
Keywords: 16S rRNA; IgA-seq; immunoglobulin A; microbiota; pre-diabetes; saliva.
Copyright © 2025 Kato-Kogoe, Tsuda, Kudo, Sakaguchi, Omori, Komori, Ohmichi, Hamada, Nakamura, Nakano, Tamaki and Ueno.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

