Mechanistic insights curcumin's anti-inflammatory in pancreatic cancer: experimental and computational evidence implicating IL1B interference via IL10RA upregulation and NLRP3/TLR3 downregulation
- PMID: 40535567
- PMCID: PMC12174075
- DOI: 10.3389/fcell.2025.1601908
Mechanistic insights curcumin's anti-inflammatory in pancreatic cancer: experimental and computational evidence implicating IL1B interference via IL10RA upregulation and NLRP3/TLR3 downregulation
Abstract
Purpose: Pancreatic cancer is a highly aggressive malignancy characterised by a complex tumour microenvironment and chronic inflammation. Studies found curcumin inhibited with inflammatory responses and tumour proliferation by interfering with production and activation of pro-inflammatory factors. This study investigated curcumin treated pancreatic cancer by modulating key targets in the inflammatory response and their signalling pathways.
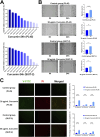
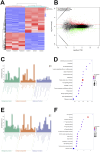
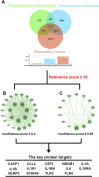
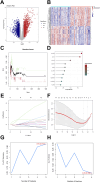
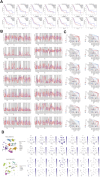
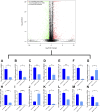
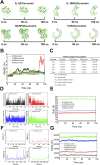
Methods: The human pancreatic cancer PL45 cells and SUIT-2 cells were utilized to establish cellular experiments, and the effects of curcumin on proliferation, apoptosis and cell migration of PL45 cells and SUIT-2 cells were detected by CCK-8, Annexin V-FITC/PI and cell scratching experiment. PL45 cells RNA from experimental and control groups was also analyzed by transcriptome sequencing. Bioinformatics screening of differential gene targets in transcriptome sequencing was performed. Gene Ontology, KEGG and Protein-protein interaction were used to analyze the differentially expressed targets at the gene level and protein level, respectively. We validated the differential gene targets by machine learning analysis of GSE28735 data, and performed survival analysis, pan-tumor analysis, immune infiltration analysis and single-cell transcriptional analysis on the differentially expressed targets. Computer simulations were utilized to verify the stability of curcumin binding to key proteins.
Results: Results of cellular experiments suggested 30 μg/mL curcumin and 50 μg/mL curcumin significantly inhibited the proliferation and growth of PL45 and SUIT-2, respectively. The transcriptome results indicated that 2,676 genes showed differential expression in curcumin-treated group compared to control group. Bioinformatics and machine learning analyses screened 14 key targets that are closely related to the inflammatory response in pancreatic cancer. Molecular dynamics showed binding free energies for IL1B/Curcumin, IL10RA/Curcumin, NLRP3/Curcumin and TLR3/Curcumin were -12.76 ± 1.41 kcal/mol, -11.42 ± 2.57 kcal/mol, -28.16 ± 3.11 kcal/mol and -12.54 ± 4.80 kcal/mol, respectively.
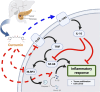
Conclusion: This research findings indicated that curcumin not only directly interfered with the activation of IL1B through blocking activation of NLRP3 by TLR3, but also upregulated expression of IL10RA to activate IL-10, thereby interfering with IL1B and its downstream signalling pathway.
Keywords: cellular experiments; computer simulation; curcumin; machine learning; pancreatic cancer; transcriptome sequencing.
Copyright © 2025 Cao, Hang, Zhang, Xia, Zhang, Men, Tian, Xia, Liao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aggarwal S., Ichikawa H., Takada Y., Sandur S. K., Shishodia S., Aggarwal B. B. (2006). Curcumin (diferuloylmethane) down-regulates expression of cell proliferation and antiapoptotic and metastatic gene products through suppression of IkappaBalpha kinase and Akt activation. Mol. Pharmacol. 69 (1), 195–206. 10.1124/mol.105.017400 - DOI - PubMed
-
- Akl L., Abd El-Hafeez A. A., Ibrahim T. M., Salem R., Marzouk H. M. M., El-Domany R. A., et al. (2022). Identification of novel piperazine-tethered phthalazines as selective CDK1 inhibitors endowed with in vitro anticancer activity toward the pancreatic cancer. Eur. J. Med. Chem. 243, 114704. 10.1016/j.ejmech.2022.114704 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

