Integrating Machine Learning Algorithms to Construct a Triaptosis-Related Prognostic Model in Melanoma
- PMID: 40535575
- PMCID: PMC12174916
- DOI: 10.2147/CMAR.S525738
Integrating Machine Learning Algorithms to Construct a Triaptosis-Related Prognostic Model in Melanoma
Abstract
Introduction: Melanoma is a highly aggressive skin cancer that accounts for a disproportionate number of skin cancer-related deaths due to early metastasis and therapy resistance. Programmed cell death (PCD), including ferroptosis and apoptosis, plays a crucial role in tumor progression and therapy response. Among these, triaptosis is a newly described form of PCD. It represents a novel mechanism of cell death with potential implications for cancer treatment. However, its role in melanoma remains largely unexplored.
Methods: We explored the role of triaptosis in melanoma by integrating single-cell and bulk RNA sequencing data. Key triaptosis-related genes and pathways were identified and incorporated into machine learning models to construct a prognostic signature. The TCGA-SKCM cohort served as the training dataset, and GEO datasets were used for validation.
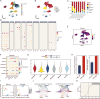
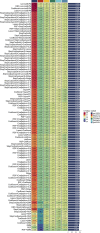
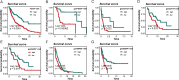
Results: A robust prognostic model based on triaptosis-associated signature (TAS) was established using the SurvivalSVM algorithm. This model showed superior predictive performance, with consistently high concordance index (C-index) values across independent validation datasets. Kaplan-Meier survival analysis indicated that high-risk patients had significantly worse overall survival than low-risk patients. The model's predictive accuracy was confirmed through receiver operating characteristic (ROC) curve analysis and principal component analysis (PCA). Moreover, immune infiltration and tumor microenvironment (TME) analyses revealed significant associations between TAS and immune cell populations.
Conclusion: Triaptosis-related gene expression patterns are closely linked with melanoma prognosis and immune infiltration. Our findings provide novel insights into triaptosis as a potential biomarker and therapeutic target, offering strategies to overcome treatment resistance in melanoma.
Keywords: cancer; cell death; melanoma; target; tumor microenvironment.
© 2025 Xie et al.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources

