Multidimensional Analysis of Twin Sets During an Intensive Week-Long Meditation Retreat: A Pilot Study
- PMID: 40535580
- PMCID: PMC12170707
- DOI: 10.1007/s12671-025-02584-x
Multidimensional Analysis of Twin Sets During an Intensive Week-Long Meditation Retreat: A Pilot Study
Abstract
Objectives: Meditation has long been known to promote health. We utilized a multidisciplinary approach to investigate the impact of mind-body interventions on the body in a twin cohort during a week-long meditation retreat.
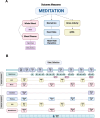
Method: This study was designed to address individual changes controlling for intersubject trait variation and explore the role of genetic background on multi-omic factors during meditation. Transcriptomic analysis was carried out from whole blood samples, while metabolomic and biochemical studies were carried out in blood plasma. Quantitative electroencephalography studies, coupled with biometric analysis and molecular studies at multiple time points, were carried out in twins meditating together and in twins separated and simultaneously either meditating or listening to a documentary.
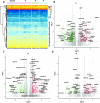
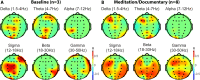
Results: Changes in gene expression, metabolites, and cytokines in blood plasma associated with specific meditative states showed patterns of change relative to the time point being assessed. Twin sets were similar in multiple domains before the start of the retreat, showed considerable divergence at the mid-point, and looked more similar by the end of the retreat. Twin pairs showed significant spectral power correlations in separate rooms and when only one twin meditated. These similarities were not observed in mismatched twin pairs. Heart rate dynamics assessments showed alignment among twin pairs, absent between unmatched pairs.
Conclusions: To our knowledge, this pilot study is novel within the twin research paradigm and is a first step toward exploring the effects of meditation in twins.
Preregistration: This study was not preregistered and was carried out under IRB protocol MED02#20211477.
Supplementary information: The online version contains supplementary material available at 10.1007/s12671-025-02584-x.
Keywords: Brain activity; Gene expression; Meditation; Metabolomics; Mind–body practices; Quantitative electroencephalography (qEEG).
© This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply 2025.
Conflict of interest statement
Conflict of interestJoe Dispenza is employed by Encephalon, Inc., which runs the meditation retreats. The remaining authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Alvarez-Lopez, M. J., Conklin, Q. A., Cosin-Tomas, M., Shields, G. S., King, B. G., Zanesco, A. P., Kaliman, P., & Saron, C. D. (2022). Changes in the expression of inflammatory and epigenetic-modulatory genes after an intensive meditation retreat. Comprehensive Psychoneuroendocrinology,11, 100152. 10.1016/j.cpnec.2022.100152 - PMC - PubMed
-
- Antoni, M. H., Lutgendorf, S. K., Blomberg, B., Carver, C. S., Lechner, S., Diaz, A., Stagl, J., Arevalo, J. M., & Cole, S. W. (2012). Cognitive-behavioral stress management reverses anxiety-related leukocyte transcriptional dynamics. Biological Psychiatry,71(4), 366–372. 10.1016/j.biopsych.2011.10.007 - PMC - PubMed
-
- Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., & Sherlock, G. (2000). Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics,25(1), 25–29. 10.1038/75556 - PMC - PubMed
LinkOut - more resources
Full Text Sources
