Antimalarial compounds exhibit variant- and cell-type-specific activity against SARS-CoV-2 isolated in Panama
- PMID: 40535766
- PMCID: PMC12175177
- DOI: 10.3389/fphar.2025.1537053
Antimalarial compounds exhibit variant- and cell-type-specific activity against SARS-CoV-2 isolated in Panama
Abstract
Background: This study evaluates the antiviral activity of antimalarial compounds against SARS-CoV-2 variants isolated in Panama (2020-2022).
Methods: For this purpose, we conducted a series of in vitro assays in two host mammalian cell systems, Vero-E6 and Calu-3 cells, to assess the antiviral activity of twenty-six antimalarials and antiviral compounds against the Delta and A2.5 variants.
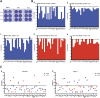
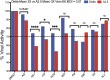
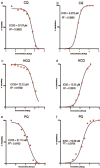
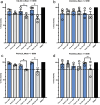
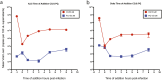
Results: In the initial screening using Vero-E6 cells, with an antiviral inhibition threshold of ≥20% and cell viability of ≥80%, chloroquine (CQ) significantly inhibited the Delta variant. Meanwhile, amodiaquine (AQ), artemisone (ASO), and ivermectin (IVM) showed activity against the A2.5 variant. In Calu-3 cells, a wider variety of compounds, including chloroquine (CQ), amodiaquine (AQ), artesunate (AS), lumefantrine (LUM), and hydroxychloroquine (HCQ), were found to be effective against the Delta variant. However, only amodiaquine (AQ) and arteether (AE) showed activity against the A2.5 variant, indicating that the response varies depending on the variant and the type of cells involved. Secondary screenings further demonstrated CQ's high inhibitory activity, with an IC50 of 6.3 μM and a selectivity index of 8, followed by HCQ, which was 1.8 times more potent against A2.5 than Delta. Time-of-addition experiments suggested that CQ and primaquine (PQ) were ineffective during the viral adsorption phase but showed a dose-dependent antiviral effect against the A2.5 variant in the early replication phase, whereas the Delta variant showed resistance.
Conclusion: This study underscores the critical role of selecting appropriate cell models for SARS-CoV-2 research, as drug efficacy varies between viral variants and host cell types.
Keywords: COVID-19; Calu-3; Panama; SARS-CoV-2 variants; Vero-E6; antimalarials; antivirals; in vitro.
Copyright © 2025 Quijada, Castillo-Bultron, Díaz, Pitti, Franco, De La Guardia, Campos, Cornejo, Núñez, Mendoza, López-Vergès, Magallon-Tejada and Obaldia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The Virus Entry Pathway Determines Sensitivity to the Antiviral Peptide TAT-I24.Viruses. 2025 Mar 23;17(4):458. doi: 10.3390/v17040458. Viruses. 2025. PMID: 40284901 Free PMC article.
-
Nanotechnology-Driven Strategy Against SARS-CoV-2: Pluronic F127-Based Nanomicelles with or Without Atazanavir Reduce Viral Replication in Calu-3 Cells.Viruses. 2025 Apr 1;17(4):518. doi: 10.3390/v17040518. Viruses. 2025. PMID: 40284961 Free PMC article.
-
Determinants of susceptibility to SARS-CoV-2 infection in murine ACE2.J Virol. 2025 Jun 17;99(6):e0054325. doi: 10.1128/jvi.00543-25. Epub 2025 May 12. J Virol. 2025. PMID: 40353671 Free PMC article.
-
Detection methods and dynamic characteristics of specific antibodies in patients with COVID-19: A review of the early literature.Heliyon. 2024 Jan 24;10(3):e24580. doi: 10.1016/j.heliyon.2024.e24580. eCollection 2024 Feb 15. Heliyon. 2024. Retraction in: Heliyon. 2025 Mar 25;11(9):e43262. doi: 10.1016/j.heliyon.2025.e43262. PMID: 38317938 Free PMC article. Retracted. Review.
-
Prevalence and odds of anxiety and depression in cutaneous malignant melanoma: a proportional meta-analysis and regression.Br J Dermatol. 2024 Jun 20;191(1):24-35. doi: 10.1093/bjd/ljae011. Br J Dermatol. 2024. PMID: 38197404
References
-
- Arshad U., Pertinez H., Box H., Tatham L., Rajoli R. K. R., Curley P., et al. (2020). Prioritization of anti-SARS-cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 108, 775–790. 10.1002/cpt.1909 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

