Genetic polymorphisms as predictors of the response of hepatocellular carcinoma patients to doxorubicin chemotherapy: a genome-wide association study
- PMID: 40535770
- PMCID: PMC12174396
- DOI: 10.3389/fphar.2025.1604473
Genetic polymorphisms as predictors of the response of hepatocellular carcinoma patients to doxorubicin chemotherapy: a genome-wide association study
Abstract
Background: Hepatocellular carcinoma (HCC), a leading cause of cancer-related mortality, is commonly treated with doxorubicin (DOX). However, its effectiveness varies significantly among patients.
Aim: The present study aimed to identify potential genetic variants affecting the response of HCC patients to DOX.
Methods: 78 patients with HCC who received DOX via transarterial chemoembolization (TACE) technology were selected. DNA was extracted from blood for genome-wide genotyping using the Applied Biosystems™ Axiom™ Precision Medicine Diversity Research™ Array. Genetic data were analysed using Axiom™ Analysis Suite software v5.2.
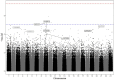
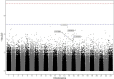
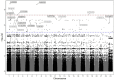
Results: Six hits in five genes [AK3 (rs378117), TRPM3 (rs1329774 and rs4745058), CDH4 (rs2427043), LINC00504 (rs76228864), and GRIN2D (rs76754767)] were associated with a risk of tumour progression, whereas variants in HPGD (rs45593131) and RC3H2 (rs2792999) were suggested as protective factors. rs8038528 in the PCSK6 gene was categorized as a low-response variant associated with an unsatisfactory reduction in α-fetoprotein (AFP) levels after DOX chemotherapy (P = 6.82 × 10-5). In contrast, three SNPs (rs1998853, rs12440990, and rs4774596) located within two genes (NPAS3 and DMXL2) were identified as predictors of good response rates to the treatment, as AFP levels were reduced by ≥ 20%. Death incidents showed associations with five SNPs that reached p ≤ 5.0 × 10-8; four of these are located within the DENND1B, LOC107986086, TMEM169, and RNF152 genes.
Conclusion: These findings support the incorporation of pharmacogenomic testing into clinical practice for HCC therapy, paving the way for customized treatment methods that may improve therapeutic efficacy and patient outcomes. Future research is needed to replicate these genetic connections.
Keywords: array genotyping; doxorubicin; genome-wide association study; hepatocellular carcinoma; transarterial chemoembolization.
Copyright © 2025 Shilbayeh, Khedr, Alshabeeb, Alsaleh, Alanizi, Abd El-Baset and Werida.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Development and validation of nomogram including mutations in angiogenesis-related genes as risk factors for HCC patients treated with TACE.Transl Oncol. 2025 Aug;58:102437. doi: 10.1016/j.tranon.2025.102437. Epub 2025 Jun 5. Transl Oncol. 2025. PMID: 40480146 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Serum CYFRA 21-1 and CK19-2G2 as Predictive Biomarkers of Response to Transarterial Chemoembolization in Hepatitis C-related Hepatocellular Carcinoma Among Egyptians: A Prospective Study.J Clin Exp Hepatol. 2025 Jan-Feb;15(1):102405. doi: 10.1016/j.jceh.2024.102405. Epub 2024 Aug 17. J Clin Exp Hepatol. 2025. PMID: 39309220
-
Prevalence and odds of anxiety and depression in cutaneous malignant melanoma: a proportional meta-analysis and regression.Br J Dermatol. 2024 Jun 20;191(1):24-35. doi: 10.1093/bjd/ljae011. Br J Dermatol. 2024. PMID: 38197404
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
References
-
- Aacr Project GENIE Consortium (2017). AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831. 10.1158/2159-8290.CD-17-0151 - DOI - PMC - PubMed
-
- Abudeif A. (2019). Epidemiology and risk factors of hepatocellular carcinoma in Egypt. Sohag Med. J. 23, 8–12. 10.21608/smj.2019.13376.1019 - DOI
-
- Balinsky D., Greengard O., Cayanis E., Head J. F. (1984). Enzyme activities and isozyme patterns in human lung tumors. Cancer Res. 44, 1058–1062. - PubMed
-
- Bessar A. A., Farag A., Monem S. M. A., Wadea F. M., Shaker S. E., Ebada M. A., et al. (2021). Transarterial chemoembolisation in patients with hepatocellular carcinoma: low-dose doxorubicin reduces post-embolisation syndrome without affecting survival-prospective interventional study. Eur. Radiol. Exp. 5, 10. 10.1186/s41747-021-00204-6 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

