Spatiotemporal Regulation of STING Activity by Linear Ubiquitination Governs Antiviral Immunity
- PMID: 40536345
- PMCID: PMC12302577
- DOI: 10.1002/advs.202417660
Spatiotemporal Regulation of STING Activity by Linear Ubiquitination Governs Antiviral Immunity
Abstract
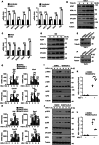
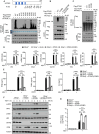
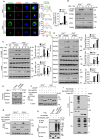
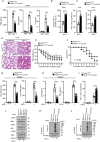
The cyclic GMP-AMP synthase (cGAS)-stimulator of interferon gene protein (STING) signaling plays a critical role in innate immunity and must be tightly regulated to maintain immune homeostasis, but the mechanism underlying the spatiotemporal regulation of this pathway remains largely elusive. Here, it is shown that during DNA viral infection, the linear ubiquitin chain assembly complex (LUBAC) and ovarian tumor deubiquitinase with linear linkage specificity (OTULIN) reversibly catalyze the linear ubiquitination of STING. At the early stage of the infection, LUBAC promotes STING linear ubiquitination to drive its trafficking from the endoplasmic reticulum (ER) to the Golgi apparatus through binding to the Sec24b subunit of the coat protein complex II (COPII) complex. Later on, OTULIN is recruited to TANK1 binding kinase 1 (TBK1)-phosphorylated STING and removes its linear ubiquitin chains, thus preventing excessive antiviral immune responses. Together, the study uncovers a linear ubiquitination-governed spatiotemporal regulatory mechanism that fine-tunes STING-driven antiviral immunity.
Keywords: HOIP; OTULIN; STING; antiviral immunity; linear ubiquitination.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
PELI2 is a negative regulator of STING signaling that is dynamically repressed during viral infection.Mol Cell. 2024 Jul 11;84(13):2423-2435.e5. doi: 10.1016/j.molcel.2024.06.001. Epub 2024 Jun 24. Mol Cell. 2024. PMID: 38917796 Free PMC article.
-
STING-ΔN, a novel splice isoform of STING, modulates innate immunity and autophagy in response to DNA virus infection.Cell Commun Signal. 2025 Jun 21;23(1):299. doi: 10.1186/s12964-025-02305-w. Cell Commun Signal. 2025. PMID: 40544261 Free PMC article.
-
Linear ubiquitination at damaged lysosomes induces local NFKB activation and controls cell survival.Autophagy. 2025 May;21(5):1075-1095. doi: 10.1080/15548627.2024.2443945. Epub 2025 Jan 2. Autophagy. 2025. PMID: 39744815
-
Stimulator of Interferon Genes (STING)-Type I Interferon Signaling: Bridging Immunity and Pain.J Integr Neurosci. 2025 Jun 23;24(6):33414. doi: 10.31083/JIN33414. J Integr Neurosci. 2025. PMID: 40613364 Review.
-
The cGAS-STING pathway in cancer immunity: dual roles, therapeutic strategies, and clinical challenges.Essays Biochem. 2025 Mar 7;69(2):EBC20253006. doi: 10.1042/EBC20253006. Essays Biochem. 2025. PMID: 40052963 Free PMC article. Review.
References
-
- Harapas C. R., Idiiatullina E., Al‐Azab M., Hrovat‐Schaale K., Reygaerts T., Steiner A., Laohamonthonkul P., Davidson S., Yu C. H., Booty L., Masters S. L., Nat. Rev. Immunol. 2022, 22, 535. - PubMed
-
- Hopfner K. P., Hornung V., Nat. Rev. Mol. Cell Biol. 2020, 21, 501. - PubMed
-
- Ogawa E., Mukai K., Saito K., Arai H., Taguchi T., Biochem. Biophys. Res. Commun. 2018, 503, 138. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
