Exploring NEK1 genetic variability in Italian amyotrophic lateral sclerosis patients
- PMID: 40536530
- PMCID: PMC12179233
- DOI: 10.1007/s00415-025-13153-6
Exploring NEK1 genetic variability in Italian amyotrophic lateral sclerosis patients
Abstract
Background: Mutations in NEK1, encoding for a serine/threonine kinase which regulates several biological processes, are associated with amyotrophic lateral sclerosis (ALS).
Methods: NEK1 was analysed by amplicon deep sequencing in a cohort of 1016 Italian sporadic and familial ALS patients previously screened for C9orf72, SOD1, TARDBP and FUS mutations.
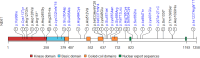
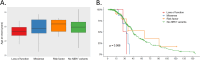
Results: We identified 28 rare NEK1 variants in 29 patients (2.85%) of whom 20/782 were sporadic (2.5%), 6/107 familial (5%) and 3/127 of unknown aetiology (2.3%). Variants were classified as pathogenic (P; n = 1), likely pathogenic (LP; n = 6 in 7 patients) and of unknown significance (VUS; n = 21) according the American College of Medical Genetics and Genomics criteria. Notably, 64% of the identified variants (18/28, including 4 LP and 14 VUS) were novel. Among the 29 patients with rare NEK1 variants, 7 (of whom 5 were familial cases) had additional variants in one of the four main ALS causative genes. Moreover, 23 patients carried the already reported NEK1 p.Arg261His risk variant (VUS) alone or in addition to SOD1 mutations (n = 1) or C9orf72 repeat expansion (n = 2) and to the NEK1 p.Asp128Val variant (n = 1). Genotype-phenotype correlation analysis showed no significant differences in age at onset or survival in NEK1 variant carriers, independently on the variant type. No flail arm phenotype, but atypical features, including sensory symptoms, were present in NEK1 carriers.
Conclusion: Our study further expands NEK1 genetic variability by identifying novel rare variants and confirming ALS oligogenic nature since 19.6% of NEK1 patients also carried mutations in one of the four main ALS-associated genes.
Keywords: NEK1; ALS; Genetic screening; IPSC; Oligogenicity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: V.S. received compensation for consulting services and/or speaking activities from AveXis, Cytokinetics, Italfarmaco, Liquidweb S.r.l., Novartis Pharma AG, Amylyx Pharmaceuticals, Biogen, and Zambon Biotech SA. He is in the Editorial Board of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, European Neurology, American Journal of Neurodegenerative Diseases, Frontiers in Neurology, and Exploration of Neuroprotective Therapy. Ethical approval: Approval for this study was obtained from the ethical committees of the participating Institutions (REB approval 2018_04_17) and studies have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Consent to participate: Informed consent was obtained from all individual participants included in the study.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

