ARID1A deficiency promotes malignant proliferation of hepatocellular carcinoma by activating HDAC7/ENO1 signaling pathway
- PMID: 40536557
- PMCID: PMC12180837
- DOI: 10.1097/HC9.0000000000000738
ARID1A deficiency promotes malignant proliferation of hepatocellular carcinoma by activating HDAC7/ENO1 signaling pathway
Abstract
Background: Epigenetic dysregulation constitutes one of the mechanisms in the pathogenesis of HCC, thereby underscoring its critical scientific importance for early diagnosis and therapeutic interventions. The chromatin remodeling factor AT-rich interaction domain 1A (ARID1A) serves as an essential epigenetic regulator and is frequently subject to inactivating mutations across various cancer types. Nevertheless, the precise molecular pathways by which ARID1A deficiency facilitates the progression of HCC remain inadequately elucidated.
Methods: RNA sequencing (RNA-seq) was employed to identify genes that were significantly upregulated following the knockdown of ARID1A. Immunohistochemistry was used to assess the expression levels of ARID1A and histone deacetylase 7 (HDAC7) in HCC samples. Chromatin immunoprecipitation and luciferase reporter assays were conducted to validate PU.1 as a positive transcriptional regulator of HDAC7. Protein immunoprecipitation coupled with mass spectrometry was used to identify potential substrates of HDAC7. A xenograft assay was performed to evaluate the therapeutic potential of HDAC7 inhibitors in ARID1A-deficient HCC.
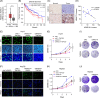
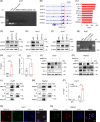
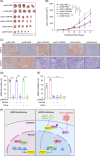
Results: Our investigation demonstrated that the loss of ARID1A in HCC cells led to an upregulation of HDAC7 expression. Mechanistic analyses revealed that ARID1A deficiency facilitated PU.1-mediated transcriptional regulation of HDAC7. Additionally, the overexpression of HDAC7 resulted in a reduced acetylation level of Enolase 1 (ENO1), thereby enhancing the malignant proliferation of HCC cells. Targeting HDAC7 effectively inhibited the growth of ARID1A-deficient tumors in tumor-bearing mice.
Conclusions: This study provides novel insights into the regulatory interplay between chromatin remodeling and acetylation modification, 2 pivotal epigenetic processes influencing tumorigenesis. It also suggests that pharmacological inhibition of HDAC7 offers a promising therapeutic strategy for treating HCC with ARID1A mutations.
Keywords: ARID1A; HCC; HDAC7; acetylation modification; chromatin remodeling.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
The authors have no conflicts to report.
Figures


Similar articles
-
mTORC1 Promotes ARID1A Degradation and Oncogenic Chromatin Remodeling in Hepatocellular Carcinoma.Cancer Res. 2021 Nov 15;81(22):5652-5665. doi: 10.1158/0008-5472.CAN-21-0206. Epub 2021 Aug 24. Cancer Res. 2021. PMID: 34429326 Free PMC article.
-
Mechanism of METTL14 regulates HBV-HCC malignant progression by mediating m6A modification of FOXP3 and thus transcriptional activation of ALDOB.J Mol Histol. 2025 Aug 8;56(4):259. doi: 10.1007/s10735-025-10551-y. J Mol Histol. 2025. PMID: 40778958
-
STOX1 Isoform A Promotes Proliferation and Progression of Hepatocellular Carcinoma by Dual Mechanisms of Transcriptionally Upregulation of Cyclin B1 and Activation of ROS-Dependent PTEN/AKT1 Signaling.Cancer Med. 2025 Jul;14(13):e70958. doi: 10.1002/cam4.70958. Cancer Med. 2025. PMID: 40567110 Free PMC article.
-
The Protective Role of miR-125b in Hepatocellular Carcinoma: Unraveling Tumor-Suppressive Mechanisms.Curr Mol Med. 2025;25(6):663-671. doi: 10.2174/0115665240304247240529074123. Curr Mol Med. 2025. PMID: 38859784 Review.
-
Molecular interplay of ARID1A in gastrointestinal cancers.Med Oncol. 2025 Aug 23;42(10):442. doi: 10.1007/s12032-025-03014-7. Med Oncol. 2025. PMID: 40848152 Review.
References
-
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Gordan JD, Kennedy EB, Abou-Alfa GK, Beg MS, Brower ST, Gade TP, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO Guideline. J Clin Oncol. 2020;38:4317–4345. - PubMed
-
- Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. - PubMed
-
- Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72:215–229. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

