Ezetimibe Engineered L14-8 Suppresses Advanced Prostate Cancer by Activating PLK1/TP53-SAT1-Induced Ferroptosis
- PMID: 40536697
- PMCID: PMC12362806
- DOI: 10.1002/advs.202504192
Ezetimibe Engineered L14-8 Suppresses Advanced Prostate Cancer by Activating PLK1/TP53-SAT1-Induced Ferroptosis
Abstract
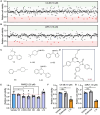
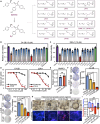
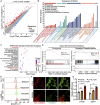
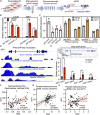
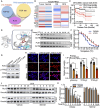
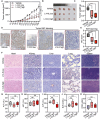
Androgen receptor signaling inhibitors (ARSIs) have demonstrated a survival benefit in metastatic prostate cancer. However, patients taking these agents inevitably acquire resistance and even develop neuroendocrine prostate cancer (NEPC), in which stage the AR signaling is inactive, and therapies are limited for these lethal cases. Therefore, developing novel treatments independent of the AR signaling pathway is urgently needed. Here it is reported that L14-8, a small molecule is derived and optimized from ezetimibe, a marketed drug primarily used for intestinal cholesterol and phytosterol absorption, significantly suppresses cell growth in advanced prostate cancer by inducing ferroptosis. Mechanistically, L14-8 binds to and promotes the ubiquitin-mediated PLK1 degradation, resulting in an increase of downstream TP53 protein phosphorylation, which is further enriched at the promoter of SAT1, a well-established ferroptosis inducer, and boosting SAT1 transcription thus triggers ferroptosis-mediated cancer cell death. Importantly, in vivo studies further demonstrate a potent anti-tumor efficacy of L14-8 without obvious toxicity. Overall, this study develops a novel small molecular engineered from ezetimibe for treating lethal prostate cancer in an AR-independent manner and provides mechanistic insights into its action by triggering PLK1-TP53-SAT1 axis-mediated ferroptosis in lethal PCa models independent of the AR signaling pathway.
Keywords: SAT1; TP53; drug design and optimization; ferroptosis; prostate cancer.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
F.Y., W.Z., Y. Z., and X.S. are co‐inventors on a patent filed by Shanghai University of Traditional Chinese Medicine that relates to the research reported in this paper. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
