Nanoparticle-Mediated CXCL12-CXCR4 Inhibition Reprograms Macrophages and Suppresses Gastric Carcinoma
- PMID: 40536774
- PMCID: PMC12376508
- DOI: 10.1002/advs.202500225
Nanoparticle-Mediated CXCL12-CXCR4 Inhibition Reprograms Macrophages and Suppresses Gastric Carcinoma
Abstract
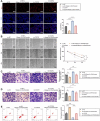
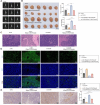
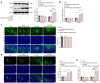
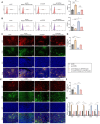
Gastric carcinoma (GC) remains a major global health challenge, requiring novel therapeutic approaches. This study investigates the efficacy of self-assembled M2pep-Cs NPs/Plerixafor nanoparticles in suppressing GC by targeting the CXCL12-CXCR4 signaling pathway and reprogramming tumor-associated macrophages (TAMs) to enhance anti-tumor immunity. The nanoparticles' physicochemical properties and biocompatibility are assessed using transmission electron microscopy, dynamic light scattering, and biological assays. A GC mouse model is established, followed by histological and immunohistochemical analyses to evaluate tumor apoptosis and proliferation. Multi-omics approaches, including transcriptomics, proteomics, and metabolomics, identify key genes and pathways affected by treatment. Flow cytometry and ELISA quantify immune activation markers; while, cell migration and invasion assays evaluate tumor suppression effects. The results demonstrate that M2pep-Cs NPs/Plerixafor effectively modulates the tumor microenvironment, suppressing GC progression by reprogramming TAMs through CXCL12-CXCR4 inhibition, enhancing immune recognition and T cell responses. This study provides mechanistic insights and highlights the potential of nanoparticle-based immunotherapy for GC, offering a promising avenue for clinical translation.
Keywords: CXCL12–CXCR4 signaling pathway; M2pep‐Cs NPs/plerixafor nanoparticles; gastric carcinoma; immunotherapy; macrophage reprogramming.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Misawa S., Denda T., Kodama S., Suzuki T., Naito Y., Kogawa T., Takada M., Suichi T., Shiosakai K., Kuwabara S., Saito G., Hino A., Imanishi S., Ureshino N., Satomi D., Tanabe Y., Hanaoka Y., Miyamoto A., Suzuki T., Naganuma A., Yanagita Y., Sekine K., Kusano F., Nakamura M., Imazeki H., BMC Cancer 2023, 23, 1098. - PMC - PubMed
-
- Griger J., Widholz S. A., Jesinghaus M., de Andrade Krätzig N., Lange S., Engleitner T., Montero J. J., Zhigalova E., Öllinger R., Suresh V., Winkler W., Lier S., Baranov O., Trozzo R., Ben Khaled N., Chakraborty S., Yu J., Konukiewitz B., Steiger K., Pfarr N., Rajput A., Sailer D., Keller G., Schirmacher P., Röcken C., Fagerstedt K. W., Mayerle J., Schmidt‐Supprian M., Schneider G., Weichert W., et al., Cancer Cell 2023, 41, 1327. - PubMed
-
- Chen Y. I‐C., Malfertheiner P., Yu H.‐T., Kuo C.‐L., Chang Y.‐Y., Meng F.‐T., Wu Y.‐X., Hsiao J.‐L., Chen M. J., Lin K. P., Wu C. Y., Lin J. T., O'Morain C., Megraud F., Lee W. C., El‐Omar E. M., Wu M. S., Liou J. M., Gastroenterology 2024, 166, 605. - PubMed
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA: Cancer J. Clin. 2021, 71, 209. - PubMed
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R. L., Soerjomataram I., Jemal A., CA: Cancer J. Clin. 2024, 74, 229. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
