ELD1 mediates photoperiodic flowering via OsCCA1 alternative splicing and interacts with phytochrome signaling in rice
- PMID: 40537466
- PMCID: PMC12179286
- DOI: 10.1038/s41467-025-60839-6
ELD1 mediates photoperiodic flowering via OsCCA1 alternative splicing and interacts with phytochrome signaling in rice
Abstract
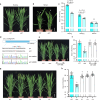
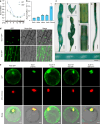
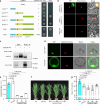
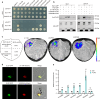
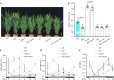
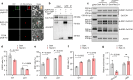
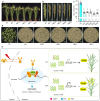
Photoperiodic flowering in plants is orchestrated by the dynamic interaction between light signals and the endogenous circadian clock, but how light signals integrate into the clock remains to be fully elucidated. Here, we identify ELD1, a CCHC-type zinc finger protein that is essential for rice embryo survival. Notably, partial loss of ELD1 function results in early flowering under long-day conditions. Further investigations demonstrate that ELD1 physically interacts with OsNKAP, an orthologue of mammal NF-κB activating protein, as well as core splicing factors to regulate the splicing profile of OsCCA1, a core oscillator of the circadian clock. Molecular and genetic evidence indicate that OsCCA1 is the primary target of ELD1 in controlling flowering time. Additionally, ELD1 interacts with photoactivated phyB, mediating red-light-regulated alternative splicing of OsCCA1. Collectively, our findings establish a molecular connection between light signaling and the circadian clock, with ELD1 modulating OsCCA1 alternative splicing to control photoperiodic flowering.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon.PLoS Genet. 2023 May 10;19(5):e1010706. doi: 10.1371/journal.pgen.1010706. eCollection 2023 May. PLoS Genet. 2023. PMID: 37163541 Free PMC article.
-
A telomere-to-telomere gapless genome reveals SlPRR1 control of circadian rhythm and photoperiodic flowering in tomato.Gigascience. 2025 Jan 6;14:giaf058. doi: 10.1093/gigascience/giaf058. Gigascience. 2025. PMID: 40601421 Free PMC article.
-
OsMADS22 interacts with OsMADS50 to regulate floral transition in rice.Biochem Biophys Res Commun. 2025 Apr 9;757:151607. doi: 10.1016/j.bbrc.2025.151607. Epub 2025 Mar 11. Biochem Biophys Res Commun. 2025. PMID: 40088677
-
A review on modeling approaches for the transcriptional regulatory network intricacies of circadian clock genes in plants.Planta. 2025 Jun 5;262(1):17. doi: 10.1007/s00425-025-04735-9. Planta. 2025. PMID: 40471439 Review.
-
The time machine: feedback loops, post-transcriptional regulation, and environmental integration in the plant circadian oscillator.Plant J. 2025 Jun;122(6):e70275. doi: 10.1111/tpj.70275. Plant J. 2025. PMID: 40538363 Review.
References
-
- Hu, Z. et al. Autophagy targets Hd1 for vacuolar degradation to regulate rice flowering. Mol. Plant15, 1137–1156 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

