Herpes simplex virus type 1 reshapes host chromatin architecture via transcription machinery hijacking
- PMID: 40537528
- PMCID: PMC12179302
- DOI: 10.1038/s41467-025-60534-6
Herpes simplex virus type 1 reshapes host chromatin architecture via transcription machinery hijacking
Abstract
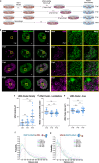
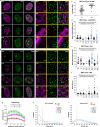
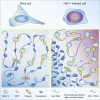
Herpes simplex virus type 1 (HSV-1) remodels the host chromatin structure and induces a host-to-virus transcriptional switch during lytic infection. We combine super-resolution imaging and chromosome-capture technologies to identify the mechanism of remodeling. We show that the host chromatin undergoes massive condensation caused by the hijacking of RNA polymerase II (RNAP II) and topoisomerase I (TOP1). In addition, HSV-1 infection results in the rearrangement of topologically associating domains and loops, although the A/B compartments are maintained in the host. The position of viral genomes and their association with RNAP II and cohesin is determined nanometrically. We reveal specific host-HSV-1 genome interactions and enrichment of upregulated human genes in the most contacting regions. Finally, TOP1 inhibition fully blocks HSV-1 infection, suggesting possible antiviral strategies. This viral mechanism of host chromatin rewiring sheds light on the role of transcription in chromatin architecture.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

