Evidence for a JAK2/STAT3 proinflammatory and vasculogenic mimicry interrelated molecular signature in adipocyte-derived mesenchymal stromal/stem cells
- PMID: 40537799
- PMCID: PMC12177984
- DOI: 10.1186/s12964-025-02298-6
Evidence for a JAK2/STAT3 proinflammatory and vasculogenic mimicry interrelated molecular signature in adipocyte-derived mesenchymal stromal/stem cells
Abstract
Background: During obesity, the excessive accumulation of fat in tissue promotes dysregulated hormonal and cytokine homeostasis that triggers chronic inflammation, which is, in part, associated with an increased incidence of some cancers. This protumoral inflammatory environment is further exacerbated through the secretome of mature adipocytes, which promotes tumor angiogenesis. Emerging studies suggest that human adipocyte-derived mesenchymal stromal/stem cells (ADMSCs) may contribute to a complementary process supporting local angiogenesis termed vasculogenic mimicry (VM). The molecular mechanisms linking ADMSCs to VM and inflammation remain poorly understood.
Methods: ADMSC 3D capillary-like structures were generated upon seeding on Cultrex. Structure analysis was performed using WIMASIS. Total RNA was extracted using TRIzol and RT-qPCR was performed to assess gene expression or screen RT2 PCR arrays. Transient gene silencing was performed by transfecting cells with a specific siRNA against STAT3. Protein lysates were harvested and used for Western blotting. Realtime cell migration was performed with the xCELLigence system.
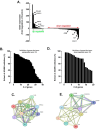
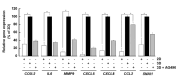
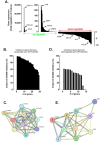
Results: Our findings revealed that in vitro priming of ADMSCs with Cultrex led to the formation of 3D capillary-like structures and the acquisition of an inflammatory molecular signature. VM-derived ADMSCs share a common proinflammatory molecular signature similar to that induced in 2D ADMSC monolayers by tumor necrosis factor (TNF)-alpha and are characterized by upregulated expression of COX2, CCL2, CCL5, CXCL5, CXCL8, IL-6, SNAI1, and MMP9. Interestingly, pharmacological inhibition or gene silencing of the JAK2/STAT3 signaling pathway reduced chemotactic cell migration, in vitro VM and the expression of proinflammatory and invasive biomarkers.
Conclusions: Overall, we provide novel evidence that inhibiting JAK2/STAT3-regulated VM can also alter the acquisition of a proinflammatory signature and prevent the contribution of ADMSCs to alternative tumor neovascularization processes.
Keywords: ADMSC; AG490; Inflammation; JAK2/STAT3; TNFα; Vasculogenic mimicry.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
A SORT1/EGFR molecular interplay as a prognostic biomarker in glioblastoma vasculogenic mimicry: implications in the transcriptional regulation of cancer stemness and drug resistance.BMC Cancer. 2025 Aug 20;25(1):1348. doi: 10.1186/s12885-025-14789-3. BMC Cancer. 2025. PMID: 40835914 Free PMC article.
-
Transcriptional regulation of CYR61 and CTGF by LM98: a synthetic YAP-TEAD inhibitor that targets in-vitro vasculogenic mimicry in glioblastoma cells.Anticancer Drugs. 2024 Sep 1;35(8):709-719. doi: 10.1097/CAD.0000000000001627. Epub 2024 Jun 18. Anticancer Drugs. 2024. PMID: 38900643 Free PMC article.
-
Tumor-expressed B7-H3 promotes vasculogenic mimicry formation rather than angiogenesis in non-small cell lung cancer.J Cancer Res Clin Oncol. 2023 Sep;149(11):8729-8741. doi: 10.1007/s00432-023-04790-3. Epub 2023 May 2. J Cancer Res Clin Oncol. 2023. PMID: 37129607 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- World Health Organization. Cancer. 2025. https://www.who.int/news-room/fact-sheets/detail/cancer
-
- Mukherjee A, Bilecz AJ, Lengyel E. The adipocyte microenvironment and cancer. Cancer Metastasis Rev. 2022;41(3):575–87. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

