Cell identity and 5-hydroxymethylcytosine
- PMID: 40537872
- PMCID: PMC12178066
- DOI: 10.1186/s13072-025-00601-w
Cell identity and 5-hydroxymethylcytosine
Abstract
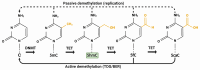
Epigenetic factors underlie cellular identity through the regulation of transcriptional networks that establish a cell's phenotype and function. Cell conversions are directed by transcription factor binding at target DNA which induce changes to identity-specific gene regulatory programs. The degree of cell plasticity is determined by the interplay of epigenetic mechanisms to create a landscape susceptible to such binding events. 5-hydroxymethylcytosine, a key intermediate during the process of DNA demethylation, is an epigenetic modification involved in controlling these epigenetic dynamics related to cell identity. Here, the role of 5-hydroxcymethylcytosine during cell identity conversions, including its relationship with other main epigenetic mechanisms, is reviewed.
Keywords: 5-hydroxymethylcytosine; Cell conversions; Cell identity; Epigenetic barriers; Epigenetic mechanisms.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Genomic context-dependent roles of 5-hydroxymethylcytosine in regulating gene expression during rice drought response.Plant J. 2025 Aug;123(4):e70436. doi: 10.1111/tpj.70436. Plant J. 2025. PMID: 40842099
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
Human brain aging is associated with dysregulation of cell type epigenetic identity.Geroscience. 2025 Jun;47(3):3759-3770. doi: 10.1007/s11357-024-01450-3. Epub 2024 Dec 27. Geroscience. 2025. PMID: 39730969 Free PMC article.
-
Epigenetic editing and epi-drugs: a combination strategy to simultaneously target KDM4 as a novel anticancer approach.Clin Epigenetics. 2025 Jun 19;17(1):105. doi: 10.1186/s13148-025-01913-0. Clin Epigenetics. 2025. PMID: 40537846 Free PMC article.
-
Decreased 5-hydroxymethylcytosine levels correlate with cancer progression and poor survival: a systematic review and meta-analysis.Oncotarget. 2017 Jan 3;8(1):1944-1952. doi: 10.18632/oncotarget.13719. Oncotarget. 2017. PMID: 27911867 Free PMC article.
Cited by
-
Systemic Neurodegeneration and Brain Aging: Multi-Omics Disintegration, Proteostatic Collapse, and Network Failure Across the CNS.Biomedicines. 2025 Aug 20;13(8):2025. doi: 10.3390/biomedicines13082025. Biomedicines. 2025. PMID: 40868276 Free PMC article. Review.
References
-
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Sci (1979). 1987;236:1237–45. - PubMed
-
- Buccitelli C, Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet. 2020;21:630–44. - PubMed
-
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comput Biol. 1999;6:281–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

