The gut virome and human health: From diversity to personalized medicine
- PMID: 40538711
- PMCID: PMC12173812
- DOI: 10.1016/j.engmic.2025.100191
The gut virome and human health: From diversity to personalized medicine
Abstract
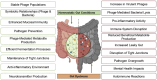
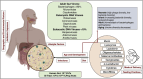
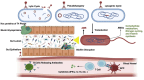
The human gut virome plays a crucial role in the gut and overall health; its diversity and regulatory functions influence bacterial populations, metabolism, and immune responses. Bacteriophages (phages) and eukaryotic viruses within the gut microbiome contribute to these processes, and recent advancements in sequencing technologies and bioinformatics have greatly expanded our understanding of the gut virome. These advances have led to the development of phage-based therapeutics, diagnostics, and artificial intelligence-driven precision medicine. The emerging field of phageomics shows promise for delivering personalized phage therapies that combat antimicrobial resistance by specifically targeting pathogenic bacteria while preserving beneficial microbes. Moreover, CRISPR-Cas systems delivered via phages have shown success in selectively targeting antibiotic resistance genes and enhancing treatment effectiveness. Phage-based diagnostics are highly sensitive in detecting bacterial pathogens, offering significant benefits for human health and zoonotic disease surveillance. This synthesis of the current knowledge highlights the pivotal role of the gut virome in regulating microbial communities and its transformative potential in personalized medicine, emphasizing its importance in advancing therapeutic and diagnostic strategies for improving health outcomes.
Keywords: Bacteriophage; CRISPR-Cas system; Dysbiosis; Fecal virome transplantation; Gut virome; Phage therapy.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Horizontal Gene Transfer and CRISPR Targeting Drive Phage-Bacterial Host Interactions and Coevolution in "Pink Berry" Marine Microbial Aggregates.Appl Environ Microbiol. 2023 Jul 26;89(7):e0017723. doi: 10.1128/aem.00177-23. Epub 2023 Jul 5. Appl Environ Microbiol. 2023. PMID: 37404190 Free PMC article.
-
An Exploration of the Relationship Between Gut Virome and Cardiovascular Disease: A Comprehensive Review.Rev Cardiovasc Med. 2025 Jun 24;26(6):36386. doi: 10.31083/RCM36386. eCollection 2025 Jun. Rev Cardiovasc Med. 2025. PMID: 40630433 Free PMC article. Review.
-
Synbiotics, prebiotics and probiotics for solid organ transplant recipients.Cochrane Database Syst Rev. 2022 Sep 20;9(9):CD014804. doi: 10.1002/14651858.CD014804.pub2. Cochrane Database Syst Rev. 2022. PMID: 36126902 Free PMC article.
-
The human phageome: niche-specific distribution of bacteriophages and their clinical implications.Appl Environ Microbiol. 2025 Jun 18;91(6):e0178824. doi: 10.1128/aem.01788-24. Epub 2025 Apr 16. Appl Environ Microbiol. 2025. PMID: 40237489 Free PMC article. Review.
-
Naturally transmitted mouse viruses highlight the heterogeneity of virus transmission dynamics in the dirty mouse model.J Virol. 2025 Jun 17;99(6):e0018725. doi: 10.1128/jvi.00187-25. Epub 2025 May 28. J Virol. 2025. PMID: 40434104 Free PMC article.
References
-
- Lathakumari R.H., Vajravelu L.K., Satheesan A., Ravi S., Thulukanam J. Antibiotics and the gut microbiome: understanding the impact on human health. Med. Microecol. 2024;20 doi: 10.1016/j.medmic.2024.100106. - DOI
Publication types
LinkOut - more resources
Full Text Sources

