Establishment and improvement of genetic manipulation tools for Fusobacterium nucleatum
- PMID: 40538714
- PMCID: PMC12173820
- DOI: 10.1016/j.engmic.2025.100192
Establishment and improvement of genetic manipulation tools for Fusobacterium nucleatum
Abstract
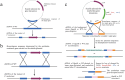
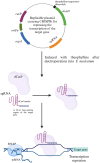
An imbalance in oral microbial homeostasis is significantly associated with the onset and progression of several systemic diseases. Fusobacterium nucleatum, a ubiquitous periodontitis-causing bacterium in the oral cavity, is frequently detected in focal sites and contributes to the pathogenesis of many extraoral diseases, including cancers, cardiovascular diseases, and adverse pregnancy outcomes (APOs). F. nucleatum is one of the few oral anaerobes that can be cultured purely in vitro and is a 'model species' for studying the impact of oral health on systemic health. The establishment and development of genetic manipulation tools for F. nucleatum and the construction of pathogenic gene-disrupted strains are important strategies for studying the pathogenicity of F. nucleatum. Here, we review the establishment and development of the genetic manipulation systems for F. nucleatum and summarize the characteristics of various genetic manipulation tools, such as suicide plasmid-based systems for gene inactivation, replicable plasmid-based systems controlling gene expression, and transposon-based random mutagenesis systems. Notably, we summarize and analyze their applications in the study of the pathogenic mechanisms of F. nucleatum. We hope to provide reference information and ideas for future research on genetic manipulation tools and the pathogenic mechanisms of F. nucleatum and other Fusobacterium species.
Keywords: Crispr interference; Fusobacterium nucleatum; Gene inactivation; Genetic manipulation tool; Natural competence; Restriction-modification system.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Addressing controversy in Fusobacterium nomenclature: what exactly does "F. nucleatum" refer to?Gut Microbes. 2025 Dec;17(1):2514797. doi: 10.1080/19490976.2025.2514797. Epub 2025 Jun 4. Gut Microbes. 2025. PMID: 40464118 Free PMC article.
-
Fusobacterium's link to colorectal neoplasia sequenced: A systematic review and future insights.World J Gastroenterol. 2017 Dec 28;23(48):8626-8650. doi: 10.3748/wjg.v23.i48.8626. World J Gastroenterol. 2017. PMID: 29358871 Free PMC article.
-
Fusobacterium nucleatum: strategies for adapting to aerobic stress.J Bacteriol. 2025 Jul 24;207(7):e0009025. doi: 10.1128/jb.00090-25. Epub 2025 Jun 6. J Bacteriol. 2025. PMID: 40476714 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Moore W.E., Moore L.V. The bacteria of periodontal diseases. Periodontology. 1994;5:66–77. . 2000. - PubMed
-
- He J., Huang W., Pan Z., Cui H., Qi G., Zhou X., Chen H. Quantitative analysis of microbiota in saliva, supragingival, and subgingival plaque of Chinese adults with chronic periodontitis. Clin. Oral Investig. 2012;16:1579–1588. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

