Transcriptome analysis reveals changes in lignin and flavonoid biosynthesis in Serendipita indica colonized Tartary buckwheat
- PMID: 40538868
- PMCID: PMC12176773
- DOI: 10.3389/fpls.2025.1595781
Transcriptome analysis reveals changes in lignin and flavonoid biosynthesis in Serendipita indica colonized Tartary buckwheat
Abstract
Introduction: Tartary buckwheat (Fagopyrum tataricum Gaertn.), classified as a food and herbal medicinal crop, offers substantial nutritional benefits but suffers from poor yields and quality. Studies indicate that Serendipita indica positively impacts Tartary buckwheat's yield and quality, yet the underlying processes remain largely unexplored.
Methods: This study aimed to examine the genetic transcript of Tartary buckwheat in both colonized and uncolonized S. indica.
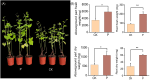
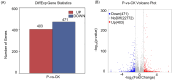
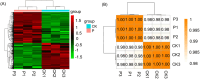
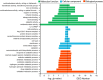
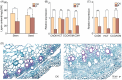
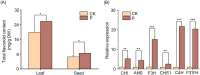
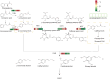
Results: It was discovered that the pathway for producing phenylpropanoids in Tartary buckwheat, both in colonized and uncolonized S. indica, both in colonized and uncolonized S. indica, was found to be enriched in KEGG (Kyoto Encyclopedia of Genes and Genomes). Genetic expression analysis of lignin and flavonoid biosynthesis pathways in colonized S. indica showed a comparison between lignin biosynthesis pathway genes in colonized S. indica and those in uncolonized S. indica in Tartary buckwheat. Research revealed a decrease in certain genes linked to lignin synthesis and an increase in others associated with flavonoid biosynthesis in both colonized and uncolonized S. indica Tartary buckwheat. Furthermore, research revealed a reduction in lignin levels in Tartary buckwheat stems and seeds both colonized and uncolonized by S. indica, in contrast to an increase in flavonoid levels in leaves and seeds of Tartary buckwheat colonized and uncolonized by the same fungi.
Discussion: Findings indicate that the process of synthesizing lignin and flavonoids could offer valuable insights into how S. indica enhances Tartary buckwheat's yield and quality.
Keywords: RNA-seq; Serendipita indica; Tartary buckwheat; flavonoid biosynthesis; lignin biosynthesis; phenylpropanoid biosynthesis.
Copyright © 2025 Wang, Zhong, Tang, Zhou, Li, Ding, Wang, Bu, Tang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Serendipita indica-dominated synthetic microbial consortia enhanced tartary buckwheat growth and improved its tolerance to drought stress.Front Microbiol. 2025 Mar 19;16:1562341. doi: 10.3389/fmicb.2025.1562341. eCollection 2025. Front Microbiol. 2025. PMID: 40177481 Free PMC article.
-
The diversity of endophytic fungi in Tartary buckwheat (Fagopyrum tataricum) and its correlation with flavonoids and phenotypic traits.Front Microbiol. 2024 Mar 14;15:1360988. doi: 10.3389/fmicb.2024.1360988. eCollection 2024. Front Microbiol. 2024. PMID: 38559356 Free PMC article.
-
Serendipita indica Promotes the Growth of Tartary Buckwheat by Stimulating Hormone Synthesis, Metabolite Production, and Increasing Systemic Resistance.J Fungi (Basel). 2023 Nov 17;9(11):1114. doi: 10.3390/jof9111114. J Fungi (Basel). 2023. PMID: 37998919 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Eliciting adverse effects data from participants in clinical trials.Cochrane Database Syst Rev. 2018 Jan 16;1(1):MR000039. doi: 10.1002/14651858.MR000039.pub2. Cochrane Database Syst Rev. 2018. PMID: 29372930 Free PMC article.
References
-
- Abdelaziz M. E., Abdelsattar M., Abdeldaym E. A., Atia M. A. M., Mahmoud A. W. M., Saad M. M., et al. (2019). Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Scientia Hortic. 256, 108532. doi: 10.1016/j.scienta.2019.05.059 - DOI
-
- Bajaj R., Huang Y., Gebrechristos S., Mikolajczyk B., Brown H., Prasad R., et al. (2018). Transcriptional responses of soybean roots to colonization with the root endophytic fungus reveals altered phenylpropanoid and secondary metabolism. Sci. Rep. 8, 10227. doi: 10.1038/s41598-018-26809-3 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

