Unveiling regulatory variants in the blood transcriptome and their association with immunity traits in pigs
- PMID: 40539042
- PMCID: PMC12177557
- DOI: 10.3389/fimmu.2025.1582982
Unveiling regulatory variants in the blood transcriptome and their association with immunity traits in pigs
Abstract
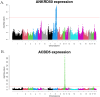
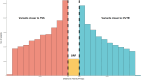
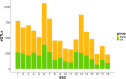
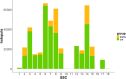
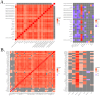
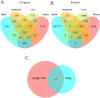
Genome wide association studies (GWAS) have been widely used to investigate the association of genetic markers with complex traits in both humans and livestock species. A particular trait of interest, when studying animal robustness and general immunocompetence, is the transcriptomic profile of blood. To identify genetic variants affecting gene expression in pig blood, we performed expression GWAS (eGWAS) in 255 animals from a commercial Duroc population between 8,499,177 imputed single nucleotide polymorphisms (SNPs) and the expression levels of 14,642 genes obtained from RNA sequencing. Out of the nearly 125 million associations tested, 23 million were found to be significant, grouped in 9,930 expression quantitative trait loci (eQTLs) associated to the expression levels of 6,051 genes. Over 36% of detected eQTLs mapped in close proximity to the genomic location of their associated gene and were classified as cis-eQTLs. Moreover, 430,694 variants were found to be associated with the expression of 10 or more different genes and were annotated as transcriptional hotspots. Among genes regulated by these hotspots, we identified genes that encode transcription factors and co-factors regulating immune responses, such as ARNT, or co-expressed genes related to immunity (CSF3R, JAK2, SOCS3, STAT5B and UBE2D1) and associated with health traits, such as phagocytic activity or haptoglobin concentration. In addition, several of the cis-regulating variants for immunity candidate genes overlapped with previously described immunity QTLs. Colocalization studies revealed putative common causal variants between the proportion of memory and helper T cells and the candidate genes CLEC12B, IGKV2D, KLRC1, KLRD1 and ZAP70. In conclusion, the associations identified in this study enable the characterization of transcriptional regulators of the pig blood transcriptome. Moreover, the colocalization between immunity QTLs and eQTLs has revealed potential causative mutations regulating immunocompetence in pigs. All these data and results contribute to shedding light on the regulatory mechanisms of blood gene expression and porcine immune regulation.
Keywords: RNA-seq; SNP; eGWAS; gene expression; immune system; lymphocytes; swine.
Copyright © 2025 Jové-Juncà, Crespo-Piazuelo, Hernández-Banqué, González-Rodríguez, Fang, Quintanilla and Ballester.
Conflict of interest statement
Author DC-P was employed by company Cuarte S.L., Spain. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

