BRD4 Signaling Maintains the Differentiated State of β Cells
- PMID: 40539402
- PMCID: PMC12412598
- DOI: 10.1002/advs.202505659
BRD4 Signaling Maintains the Differentiated State of β Cells
Abstract
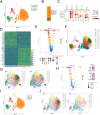
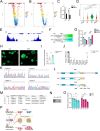
In diabetes, pancreatic β cells degenerate from their mature differentiated state to a dedifferentiated state. BRD4 plays a pivotal role during embryogenesis and cancer development, but its function in modulating β-cell differentiation remains unknown. In this study, multiple models including calorie restriction db/db mouse, long-term and acute conditional knockout mouse, and human islet organoids are adopted to assess BRD4 function in β cells. Two hundred twenty-two young patients with diabetes are also recruited for whole exome sequencing (WES) to screen for BRD4 mutations. This study shows that BRD4 expression is significantly reduced in human diabetic β cells while significantly increased after calorie restriction in the diabetic mouse. β cell differentiation is impaired after long-term and acute Brd4 knockout. BRD4 knockdown in human islet organoids results in the loss of differentiation and reduction of insulin synthesis. It is found that p.R749C can significantly affect BRD4 signaling and might play roles in diabetes development in patients. This study also shows that ATF5 is a direct target of the BRD4 pathway in β cells. Targeting BRD4-mediated regulatory networks may hold promise for developing novel therapeutic strategies to maintain the differentiated state of β cells.
Keywords: ATF5; BRD4; diabetes; differentiation; β cell.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Vetere A., Choudhary A., Burns S. M., Wagner B. K., Nat. Rev. Drug Discovery 2014, 13, 278. - PubMed
MeSH terms
Substances
Grants and funding
- 2023ZD0507700/Noncommunicable Chronic Diseases-National Science and Technology Major Project
- 2022YFA1004800/National Key Research and Development Program of China
- 2023YFA1801104/National Key Research and Development Program of China
- 82270845/National Natural Science Foundation of China
- 82070868/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous
