CACHE Challenge #2: Targeting the RNA Site of the SARS-CoV-2 Helicase Nsp13
- PMID: 40539604
- PMCID: PMC12323591
- DOI: 10.1021/acs.jcim.5c00535
CACHE Challenge #2: Targeting the RNA Site of the SARS-CoV-2 Helicase Nsp13
Abstract
A critical assessment of computational hit-finding experiments (CACHE) challenge was conducted to predict ligands for the SARS-CoV-2 Nsp13 helicase RNA binding site, a highly conserved COVID-19 target. Twenty-three participating teams comprised of computational chemists and data scientists used protein structure and data from fragment-screening paired with advanced computational and machine learning methods to each predict up to 100 inhibitory ligands. Across all teams, 1957 compounds were predicted and were subsequently procured from commercial catalogs for biophysical assays. Of these compounds, 0.7% were confirmed to bind to Nsp13 in a surface plasmon resonance assay. The six best-performing computational workflows used fragment growing, active learning, or conventional virtual screening with and without complementary deep-learning scoring functions. Follow-up functional assays resulted in identification of two compound scaffolds that bound Nsp13 with a Kd below 10 μM and inhibited in vitro helicase activity. Overall, CACHE #2 participants were successful in identifying hit compound scaffolds targeting Nsp13, a central component of the coronavirus replication-transcription complex. Computational design strategies recurrently successful across the first two CACHE challenges include linking or growing docked or crystallized fragments and docking small and diverse libraries to train ultrafast machine-learning models. The CACHE #2 competition reveals how crowd-sourcing ligand prediction efforts using a distinct array of approaches followed with critical biophysical assays can result in novel lead compounds to advance drug discovery efforts.
Conflict of interest statement
Disclosures
The authors declare the following competing financial interest(s): U.L, Y.W and L.W are full-time employees of Boehringer Ingelheim, Y.S and A.H are full time employees of UCB and may also be stockholders.
Figures



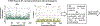



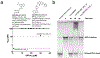
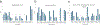
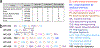

References
-
- Ackloo S; Al-Awar R; Amaro RE; Arrowsmith CH; Azevedo H; Batey RA; Bengio Y; Betz UAK; Bologa CG; Chodera JD; Cornell WD; Dunham I; Ecker GF; Edfeldt K; Edwards AM; Gilson MK; Gordijo CR; Hessler G; Hillisch A; Hogner A; Irwin JJ; Jansen JM; Kuhn D; Leach AR; Lee AA; Lessel U; Morgan MR; Moult J; Muegge I; Oprea TI; Perry BG; Riley P; Rousseaux SAL; Saikatendu KS; Santhakumar V; Schapira M; Scholten C; Todd MH; Vedadi M; Volkamer A; Willson TM CACHE (Critical Assessment of Computational Hit-Finding Experiments): A Public-Private Partnership Benchmarking Initiative to Enable the Development of Computational Methods for Hit-Finding. Nat Rev Chem 2022, 6 (4), 287–295. 10.1038/s41570-022-00363-z. - DOI - PMC - PubMed
-
- Li F; Ackloo S; Arrowsmith CH; Ban F; Barden CJ; Beck H; Beránek J; Berenger F; Bolotokova A; Bret G; Breznik M; Carosati E; Chau I; Chen Y; Cherkasov A; Corte DD; Denzinger K; Dong A; Draga S; Dunn I; Edfeldt K; Edwards A; Eguida M; Eisenhuth P; Friedrich L; Fuerll A; Gardiner SS; Gentile F; Ghiabi P; Gibson E; Glavatskikh M; Gorgulla C; Guenther J; Gunnarsson A; Gusev F; Gutkin E; Halabelian L; Harding RJ; Hillisch A; Hoffer L; Hogner A; Houliston S; Irwin JJ; Isayev O; Ivanova A; Jacquemard C; Jarrett AJ; Jensen JH; Kireev D; Kleber J; Koby SB; Koes D; Kumar A; Kurnikova MG; Kutlushina A; Lessel U; Liessmann F; Liu S; Lu W; Meiler J; Mettu A; Minibaeva G; Moretti R; Morris CJ; Narangoda C; Noonan T; Obendorf L; Pach S; Pandit A; Perveen S; Poda G; Polishchuk P; Puls K; Pütter V; Rognan D; Roskams-Edris D; Schindler C; Sindt F; Spiwok V; Steinmann C; Stevens RL; Talagayev V; Tingey D; Vu O; Walters WP; Wang X; Wang Z; Wolber G; Wolf CA; Wortmann L; Zeng H; Zepeda CA; Zhang KYJ; Zhang J; Zheng S; Schapira M CACHE Challenge #1: Targeting the WDR Domain of LRRK2, A Parkinson’s Disease Associated Protein. J Chem Inf Model 2024, 64 (22), 8521–8536. 10.1021/acs.jcim.4c01267. - DOI - PubMed
-
- Newman JA; Douangamath A; Yadzani S; Yosaatmadja Y; Aimon A; Brandão-Neto J; Dunnett L; Gorrie-Stone T; Skyner R; Fearon D; Schapira M; von Delft F; Gileadi O Structure, Mechanism and Crystallographic Fragment Screening of the SARS-CoV-2 NSP13 Helicase. Nat Commun 2021, 12 (1), 4848. 10.1038/s41467-021-25166-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

