NPR1 Promotes Lipid Droplet Lipolysis to Enhance Mitochondrial Oxidative Phosphorylation and Fuel Gastric Cancer Metastasis
- PMID: 40539884
- PMCID: PMC12499470
- DOI: 10.1002/advs.202503233
NPR1 Promotes Lipid Droplet Lipolysis to Enhance Mitochondrial Oxidative Phosphorylation and Fuel Gastric Cancer Metastasis
Abstract
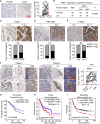
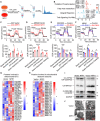
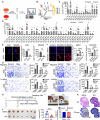
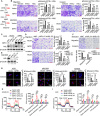
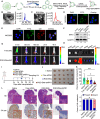
Metabolic reprogramming driven by oncogenes plays a critical role in promoting and sustaining multiple steps of gastric cancer metastasis. However, the key metabolic driver of metastasis that can lead to the development of targeted therapies for preventing and treating metastatic gastric cancer remains elusive. Here, it is identified that the transmembrane guanylate cyclase, natriuretic peptide receptor 1 (NPR1), promoted gastric cancer lymph node metastasis by activating lipid droplet lipolysis and enhancing mitochondrial oxidative phosphorylation (OXPHOS). Clinical analysis reveals that elevated NPR1 protein level is correlated with increased lymph node metastasis and shorter patient survival. Functionally, NPR1 induced lipolysis of stored lipid droplets, releasing bioavailable fatty acids that are imported into mitochondria to upregulate OXPHOS, thus fueling the energy required for the metastasis of gastric cancer cells. Mechanistically, NPR1 activates protein kinase cGMP-dependent 1 (PRKG1 or PKG), which directly bound to and activated hormone-sensitive lipase (HSL) by phosphorylation at residues Ser855 and Ser951, thereby increasing lipolysis. Furthermore, targeted delivery of NPR1 siRNA using engineered exosome mimetics effectively suppressed gastric cancer metastasis. Taken together, these findings elucidate the NPR1-driven metabolic mechanism underlying gastric cancer metastasis and suggest NPR1 as a promising therapeutic target for patients with metastatic gastric cancer.
Keywords: engineered cell membrane‐derived exosome mimetics; gastric cancer; lipid droplet; lipolysis; lymph node metastasis; mitochondrial oxidative phosphorylation; natriuretic peptide receptor 1.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Ca‐Cancer J. Clin. 2021, 71, 209. - PubMed
MeSH terms
Substances
Grants and funding
- 82172637/National Natural Science Foundation of China
- 82303240/National Natural Science Foundation of China
- 82373026/National Natural Science Foundation of China
- 32371472/National Natural Science Foundation of China
- 82000206/National Natural Science Foundation of China
- ZSQYJZPI202010/Research start-up fund of part-time PI, SAHSYSU
- 2021B1212040006/Guangdong Provincial Key Laboratory of Digestive Cancer Research
- 0030251006/Guangdong Provincial Key Laboratory of Digestive Cancer Research
- ZSQYRSFPD0062/Research Start-up Fund of Post-doctoral of SAHSYSU
- 2023A0505050120/Guangdong International Science and Technology Cooperation Project
- GJHZ20220913142401003/Shenzhen International Cooperative Research Project
- 2023A1515011962/Shenzhen International Cooperative Research Project
- Natural Science Foundation of Guangdong Province
- R08044/Kelin New Star of the First Affiliated Hospital of Sun Yat-Sen University
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
