Deciphering the Regulatory Networks of the Migrasome-Associated Cell Subpopulation in Heterotopic Ossification via Multi-Omics Analysis
- PMID: 40540299
- PMCID: PMC12180572
- DOI: 10.1096/fj.202500965R
Deciphering the Regulatory Networks of the Migrasome-Associated Cell Subpopulation in Heterotopic Ossification via Multi-Omics Analysis
Abstract
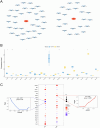
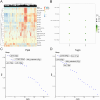
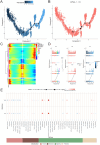
Heterotopic ossification (HO) is a pathological process where bone forms in extraskeletal tissues, often occurring as a complication of tissue repair following injury. This condition can lead to movement limitations, pain, and functional impairment. However, the underlying pathomechanisms remain poorly understood. This study aims to elucidate key biomolecular networks involved in HO through a comprehensive multi-omics analysis. Single-cell, bulk, and spatial transcriptome datasets were obtained from the Gene Expression Omnibus (GEO) database. Migrasome score analysis identified a critical cell subtype associated with HO. Key genes were identified through high-dimensional weighted gene co-expression network analysis (hdWGCNA), machine learning, and dataset validation from clinical samples. Then we analyzed immune infiltration, microRNA (miRNA) networks, co-expression networks, transcription factor (TF) regulatory networks, and signaling pathways to investigate potential regulatory mechanisms of HO. Spatial transcriptomics revealed the spatial patterns of cell subpopulation distribution and key molecule expression. Experimental validation further confirmed the expression patterns of key molecules in HO. As a result, we identified mesenchymal lineage cells (MLin) as the key migrasome-associated cell subtype and determined peptidylprolyl isomerase B (Ppib) and transgelin (Tagln) as the key molecules. We constructed a regulatory network of these biomolecules and clarified their spatial distribution. Notably, the expression of Ppib and Tagln is temporally correlated with HO progression. Collectively, the identification of Ppib and Tagln, along with the construction of key biomolecular networks, facilitates the discovery of novel biomarkers for HO, offering promising potential for the development of preventive and therapeutic strategies.
Keywords: hdWGCNA; heterotopic ossification; machine learning; migrasome; single‐cell RNA sequencing; spatial transcriptomics.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Xu R., Hu J., Zhou X., and Yang Y., “Heterotopic Ossification: Mechanistic Insights and Clinical Challenges,” Bone 109 (2018): 134–142. - PubMed
-
- Hoyt B. W., Pavey G. J., Potter B. K., and Forsberg J. A., “Heterotopic Ossification and Lessons Learned From Fifteen Years at War: A Review of Therapy, Novel Research, and Future Directions for Military and Civilian Orthopaedic Trauma,” Bone 109 (2018): 3–11. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

