Transcriptomic Differences Between Immortalized Oral and Skin Keratinocytes
- PMID: 40540308
- PMCID: PMC12180575
- DOI: 10.1096/fj.202500374R
Transcriptomic Differences Between Immortalized Oral and Skin Keratinocytes
Abstract
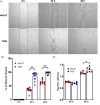
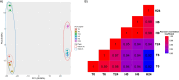
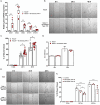
Compared to skin wounds, oral mucosal wounds heal quicker with less inflammation, faster re-epithelialization, and minimal scarring. Site-specific keratinocytes may be one differentiating factor. This study used immortalized skin and oral keratinocytes (HaCaT and TIGK), which maintain fidelity to their primary cell counterpart, to examine functional and transcriptional differences that might contribute to the differential wound healing at the two sites. Oral keratinocytes were found to have an enhanced migratory and proliferative capacity. To examine the transcriptomic differences, we generated an mRNA-sequencing gene expression dataset utilizing HaCaT and TIGK. Differentially expressed genes (DEGs) were identified between HaCaT and TIGK at baseline and throughout in vitro healing. DEGs in HaCaT and TIGK following injury were also identified when compared to each respective cell type's unwounded gene expression levels. Gene set enrichment analyses were performed to understand the biological significance of the DEGs. Processes related to interferon (IFN) signaling were uniquely enriched in TIGK. TIGK also exhibited a faster transcriptional response to injury and differential expression of integrins and matrix metalloproteinases (MMPs). When grown on extracellular matrix (ECM) proteins, TIGK retained its enhanced migratory capacity over HaCaT. Lastly, TIGK displayed a post-injury secretome that promoted keratinocyte migration. Our comparative analyses suggest that specific transcriptomic differences between oral and skin keratinocytes at unwounded baseline and in response to injury may underlie the distinct wound healing phenotypes observed in these two tissues. This work also provides a new resource of HaCaT and TIGK gene expression data that can be used for future analyses.
Keywords: keratinocytes; mouth mucosa; skin; transcription factors; wound healing.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Exome and transcriptome analysis link calcium channel pathway aberrations to botulinum toxin A resistance in Hailey-Hailey disease.Br J Dermatol. 2025 Jun 20;193(1):147-156. doi: 10.1093/bjd/ljaf112. Br J Dermatol. 2025. PMID: 40317184
-
Topical antimicrobial agents for treating foot ulcers in people with diabetes.Cochrane Database Syst Rev. 2017 Jun 14;6(6):CD011038. doi: 10.1002/14651858.CD011038.pub2. Cochrane Database Syst Rev. 2017. PMID: 28613416 Free PMC article.
-
SPRR1B+ keratinocytes prime oral mucosa for rapid wound healing via STAT3 activation.Commun Biol. 2024 Sep 16;7(1):1155. doi: 10.1038/s42003-024-06864-5. Commun Biol. 2024. PMID: 39300285 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Szpaderska A. M., Zuckerman J. D., and DiPietro L. A., “Differential Injury Responses in Oral Mucosal and Cutaneous Wounds,” Journal of Dental Research 82 (2003): 621–626. - PubMed
-
- Szpaderska A. M., Walsh C. G., Steinberg M. J., and DiPietro L. A., “Distinct Patterns of Angiogenesis in Oral and Skin Wounds,” Journal of Dental Research 84 (2005): 309–314. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

