Angiopoietin-like protein 2 mediates vasculopathy-driven fibrogenesis in a mouse model of systemic sclerosis
- PMID: 40540411
- PMCID: PMC12321390
- DOI: 10.1172/JCI177123
Angiopoietin-like protein 2 mediates vasculopathy-driven fibrogenesis in a mouse model of systemic sclerosis
Abstract
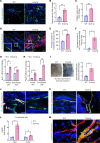
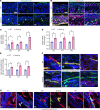
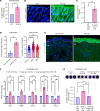
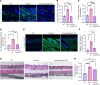
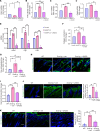
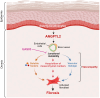
Vasculopathy is a common hallmark of various fibrotic disorders, including systemic sclerosis (SSc), yet its underlying etiology and contribution to fibrogenesis remain ill defined. In SSc, the vasculopathy typically precedes the onset of fibrosis, and we observed that this phenomenon is recapitulated in the Snail transgenic mouse model of SSc. The vascular anomalies manifest as deformed vessels, endothelial cell dysfunction, and vascular leakage. Our investigation into the underlying mechanism of this phenotype revealed that angiopoietin-like protein 2 (ANGPTL2), secreted by the Snail transgenic keratinocytes, is a principal driver of fibrotic vasculopathy. In endothelial cells, ANGPTL2 upregulates profibrotic genes, downregulates various junctional proteins, and prompts the acquisition of mesenchymal characteristics. Inhibiting endothelial cell junctional instability and, consequently, vascular leakage with a synthetic analog of the microbial metabolite Urolithin A (UAS03) effectively mitigated the vasculopathy and inhibited fibrogenesis. Thus, ANGPTL2 is a promising early biomarker of the disease, and inhibiting the vasculopathy-inducing effects of this protein with agents such as UAS03 presents an appealing therapeutic avenue to reduce disease severity. These insights hold the potential to revolutionize the approach to treatment of fibrotic diseases by targeting vascular defects.
Keywords: Cell biology; Dermatology; Endothelial cells; Fibrosis; Skin; Vascular biology.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

