SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion
- PMID: 40540531
- PMCID: PMC12208438
- DOI: 10.1371/journal.ppat.1013295
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion
Abstract
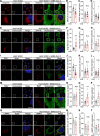
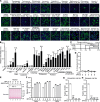
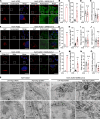
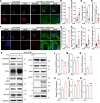
The COVID-19 pandemic is caused by the enveloped virus SARS-CoV-2. Despite extensive investigation, the molecular mechanisms for its assembly and secretion remain largely elusive. Here, we show that SARS-CoV-2 infection induces global alterations of the host endomembrane system, including dramatic Golgi fragmentation. SARS-CoV-2 virions are enriched in the fragmented Golgi. Blocking endoplasmic reticulum (ER) to Golgi trafficking dramatically inhibits SARS-CoV-2 assembly and secretion without reducing viral genome replication. Significantly, SARS-CoV-2 infection down-regulates GRASP55 but up-regulates TGN46 protein levels. Surprisingly, GRASP55 expression reduces both viral secretion and spike number on each virion without affecting viral entry, while GRASP55 depletion displays opposite effects. In contrast, TGN46 depletion only inhibits viral secretion without affecting spike incorporation into virions. Taken together, we show that SARS-CoV-2 alters Golgi structure and function to modulate viral assembly and secretion, highlighting the Golgi as a potential therapeutic target for blocking SARS-CoV-2 infection.
Copyright: © 2025 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Update of
-
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion.bioRxiv [Preprint]. 2024 Mar 15:2022.03.04.483074. doi: 10.1101/2022.03.04.483074. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Jun 20;21(6):e1013295. doi: 10.1371/journal.ppat.1013295. PMID: 35291301 Free PMC article. Updated. Preprint.
Similar articles
-
COP I vesicles facilitate classical swine fever virus proliferation by transporting fatty acid synthase from the Golgi apparatus to the endoplasmic reticulum.J Virol. 2025 Jul 22;99(7):e0030525. doi: 10.1128/jvi.00305-25. Epub 2025 Jun 3. J Virol. 2025. PMID: 40459259 Free PMC article.
-
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion.bioRxiv [Preprint]. 2024 Mar 15:2022.03.04.483074. doi: 10.1101/2022.03.04.483074. bioRxiv. 2024. Update in: PLoS Pathog. 2025 Jun 20;21(6):e1013295. doi: 10.1371/journal.ppat.1013295. PMID: 35291301 Free PMC article. Updated. Preprint.
-
Host protein ARF1 is a proviral factor for SARS-CoV-2 and a candidate broad-spectrum therapeutic target.Nat Commun. 2025 Jul 9;16(1):6326. doi: 10.1038/s41467-025-61431-8. Nat Commun. 2025. PMID: 40634337 Free PMC article.
-
SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis.Virology. 2025 Sep;610:110607. doi: 10.1016/j.virol.2025.110607. Epub 2025 Jun 12. Virology. 2025. PMID: 40544703 Review.
-
Non-pharmacological measures implemented in the setting of long-term care facilities to prevent SARS-CoV-2 infections and their consequences: a rapid review.Cochrane Database Syst Rev. 2021 Sep 15;9(9):CD015085. doi: 10.1002/14651858.CD015085.pub2. Cochrane Database Syst Rev. 2021. PMID: 34523727 Free PMC article.
Cited by
-
The SARS-CoV-2 envelope PDZ binding motif acts as a virulence factor disrupting host's epithelial cell-cell junctions.Cell Mol Biol Lett. 2025 Jul 11;30(1):80. doi: 10.1186/s11658-025-00758-y. Cell Mol Biol Lett. 2025. PMID: 40646464 Free PMC article.
-
COP I vesicles facilitate classical swine fever virus proliferation by transporting fatty acid synthase from the Golgi apparatus to the endoplasmic reticulum.J Virol. 2025 Jul 22;99(7):e0030525. doi: 10.1128/jvi.00305-25. Epub 2025 Jun 3. J Virol. 2025. PMID: 40459259 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

