Senataxin and DNA-PKcs redundantly promote non-homologous end joining repair of DNA double strand breaks during V(D)J recombination
- PMID: 40540553
- PMCID: PMC12180482
- DOI: 10.1126/sciadv.ads5272
Senataxin and DNA-PKcs redundantly promote non-homologous end joining repair of DNA double strand breaks during V(D)J recombination
Abstract
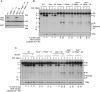
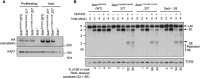
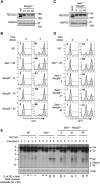
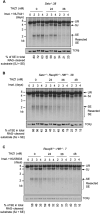
Nonhomologous end joining (NHEJ) is required for repairing DNA double strand breaks (DSBs) generated by the RAG endonuclease during lymphocyte antigen receptor gene assembly by V(D)J recombination. The ataxia telangiectasia-mutated (ATM) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) kinases regulate functionally redundant pathways required for NHEJ. Here, we report that loss of the senataxin helicase leads to a strong defect in RAG DSB repair upon inactivation of DNA-PKcs. The NHEJ function of senataxin is redundant with the RECQL5 helicase and the HLTF translocase and is epistatic with ATM. Co-inactivation of ATM, RECQL5, and HLTF results in an NHEJ defect similar to that from the combined deficiency of DNA-PKcs and senataxin or losing senataxin, RECQL5, and HLTF. These data suggest that ATM and DNA-PKcs regulate the functions of senataxin and RECQL5/HLTF, respectively, to provide redundant support for NHEJ.
Figures






Update of
-
Senataxin and DNA-PKcs Redundantly Promote Non-Homologous End Joining Repair of DNA Double Strand Breaks During V(D)J Recombination.bioRxiv [Preprint]. 2024 Sep 26:2024.09.25.615014. doi: 10.1101/2024.09.25.615014. bioRxiv. 2024. Update in: Sci Adv. 2025 Jun 20;11(25):eads5272. doi: 10.1126/sciadv.ads5272. PMID: 39386666 Free PMC article. Updated. Preprint.
Similar articles
-
Senataxin and DNA-PKcs Redundantly Promote Non-Homologous End Joining Repair of DNA Double Strand Breaks During V(D)J Recombination.bioRxiv [Preprint]. 2024 Sep 26:2024.09.25.615014. doi: 10.1101/2024.09.25.615014. bioRxiv. 2024. Update in: Sci Adv. 2025 Jun 20;11(25):eads5272. doi: 10.1126/sciadv.ads5272. PMID: 39386666 Free PMC article. Updated. Preprint.
-
Functional intersection of ATM and DNA-dependent protein kinase catalytic subunit in coding end joining during V(D)J recombination.Mol Cell Biol. 2013 Sep;33(18):3568-79. doi: 10.1128/MCB.00308-13. Epub 2013 Jul 8. Mol Cell Biol. 2013. PMID: 23836881 Free PMC article.
-
Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):2234-9. doi: 10.1073/pnas.1222573110. Epub 2013 Jan 23. Proc Natl Acad Sci U S A. 2013. PMID: 23345432 Free PMC article.
-
Functional overlaps between XLF and the ATM-dependent DNA double strand break response.DNA Repair (Amst). 2014 Apr;16:11-22. doi: 10.1016/j.dnarep.2014.01.010. Epub 2014 Feb 20. DNA Repair (Amst). 2014. PMID: 24674624 Free PMC article. Review.
-
DNA-Dependent Protein Kinase Catalytic Subunit: The Sensor for DNA Double-Strand Breaks Structurally and Functionally Related to Ataxia Telangiectasia Mutated.Genes (Basel). 2021 Jul 27;12(8):1143. doi: 10.3390/genes12081143. Genes (Basel). 2021. PMID: 34440313 Free PMC article. Review.
References
-
- Bassing C. H., Swat W., Alt F. W., The mechanism and regulation of chromosomal V(D)J recombination. Cell 109, S45–S55 (2002). - PubMed
-
- Lieber M. R., Ma Y., Pannicke U., Schwarz K., The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair 3, 817–826 (2004). - PubMed
-
- Rooney S., Chaudhuri J., Alt F. W., The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200, 115–131 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

