TGFβ-dependent signaling drives tumor growth and aberrant extracellular matrix dynamics in NF1-associated plexiform neurofibroma
- PMID: 40540562
- PMCID: PMC12180511
- DOI: 10.1126/sciadv.adu0772
TGFβ-dependent signaling drives tumor growth and aberrant extracellular matrix dynamics in NF1-associated plexiform neurofibroma
Abstract
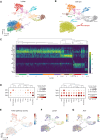
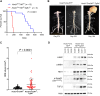
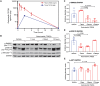
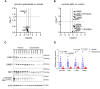
Plexiform neurofibromas (PNFs) are benign tumors of the peripheral nervous system that represent a major source of morbidity in neurofibromatosis type 1 (NF1). A substantial proportion of patients do not respond to current therapies or experience intolerable side effects. Transcriptomic characterization of murine and human PNF at bulk and single-cell resolution identified transforming growth factor-β (TGFβ) signaling as a key upstream regulator, driving aberrant basement membrane (BM) protein production by neoplastic Schwann cells and Fbs. Conditional TGFβ1 overexpression in Nf1-deficient Schwann cells driven by Hoxb7-Cre promoted PNF growth and malignant transformation in vivo. Conversely, pharmacologic inhibition of the type I TGFβ receptor (TGFβRI) reduced PNF tumor burden in Nf1 mutant mice. Proteomic characterization of the extracellular matrix (ECM) showed reduced BM proteins upon TGFβRI inhibition. These findings implicate TGFβ as a potential therapeutic target in PNF and provide insights into the role of TGFβ signaling in orchestrating ECM dynamics in the PNF microenvironment.
Figures



References
-
- Peltonen J., Penttinen R., Larjava H., Aho H. J., Collagens in neurofibromas and neurofibroma cell cultures. Ann. N. Y. Acad. Sci. 486, 260–270 (1986). - PubMed
-
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O’Connell P., Cawthon R. M., Innis M. A., McCormick F., The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63, 843–849 (1990). - PubMed
-
- Viskochil D., Buchberg A. M., Xu G., Cawthon R. M., Stevens J., Wolff R. K., Culver M., Carey J. C., Copeland N. G., Jenkins N. A., White R., O’Connell P., Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 62, 187–192 (1990). - PubMed
-
- Gross A. M., Wolters P. L., Dombi E., Baldwin A., Whitcomb P., Fisher M. J., Weiss B., Kim A., Bornhorst M., Shah A. C., Martin S., Roderick M. C., Pichard D. C., Carbonell A., Paul S. M., Therrien J., Kapustina O., Heisey K., Clapp D. W., Zhang C., Peer C. J., Figg W. D., Smith M., Glod J., Blakeley J. O., Steinberg S. M., Venzon D. J., Doyle L. A., Widemann B. C., Selumetinib in children with inoperable plexiform neurofibromas. N. Engl. J. Med. 382, 1430–1442 (2020). - PMC - PubMed
-
- McCowage G. B., Mueller S., Pratilas C. A., Hargrave D. R., Moertel C. L., Whitlock J., Fox E., Hingorani P., Russo M. W., Dasgupta K., Tseng L., Mookerjee B., Geoerger B., Trametinib in pediatric patients with neurofibromatosis type 1 (NF-1)–associated plexiform neurofibroma: A phase I/IIa study. J. Clin. Oncol. 36, 10504–10504 (2018).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

