Regulation of MORC-1 is key to the CSR-1-mediated germline gene licensing mechanism in C. elegans
- PMID: 40540580
- PMCID: PMC12180489
- DOI: 10.1126/sciadv.ado4170
Regulation of MORC-1 is key to the CSR-1-mediated germline gene licensing mechanism in C. elegans
Abstract
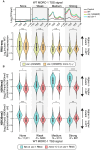
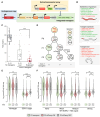
The Argonaute CSR-1 is essential for germline development in C. elegans. Loss of CSR-1 leads to the down-regulation of thousands of germline-expressed genes, supporting a model in which CSR-1 "licenses" gene expression via a poorly understood mechanism. In contrast, a small subset of genes is up-regulated in csr-1 mutants, including morc-1, which encodes a conserved GHKL-type ATPase. We show that morc-1 is overexpressed in csr-1 mutants and accumulates over CSR-1 licensed targets, coinciding with aberrant gain of H3K9me3, reduced H3K36me3, and transcriptional repression. Notably, loss of morc-1 fully rescues these chromatin defects and partially restores gene expression and fertility in csr-1 mutants. Conversely, ectopic overexpression of MORC-1 in the wild-type germ line is sufficient to repress CSR-1 licensed targets and severely compromise fertility. These findings support a model in which CSR-1 prevents MORC-1 overexpression and consequent misregulation of CSR-1 licensed genes.
Figures



References
-
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C.-C. G., Tolia N. H., Joshua-Tor L., Mitani S., Simard M. J., Mello C. C., Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757 (2006). - PubMed
-
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., van Wolfswinkel J. C., Chaves D. A., Shirayama M., Mitani S., Ketting R. F., Conte D., Mello C. C., The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134 (2009). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

