The glucocorticoid receptor is affected by its target ZBTB16 in a dissociated manner
- PMID: 40540650
- PMCID: PMC12231182
- DOI: 10.1530/JOE-24-0283
The glucocorticoid receptor is affected by its target ZBTB16 in a dissociated manner
Abstract
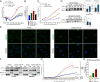
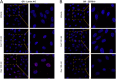
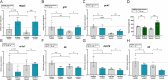
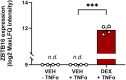
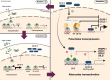
The glucocorticoid receptor (GR) mediates many activating and repressive effects of glucocorticoids in multiple contexts. Glucocorticoids can robustly induce the transcriptionally active protein Zinc finger and BTB domain containing 16 (ZBTB16). We evaluated how cortisol-induced ZBTB16, in turn, affects various GR-mediated actions in human cells and in zebrafish. We found that prevention of ZBTB16 induction led to potentiated GR-dependent effects on the human endothelial cell barrier and blood glucose levels in zebrafish larvae. In contrast, zbtb16 functional knockout abolished the GR-dependent effects on the inflammatory response in zebrafish larvae. At the mRNA level, zbtb16 knockdown potentiated transactivation and attenuated transrepression in a subset of GR target genes. Finally, ZBTB16 protein was strongly induced by dexamethasone in fibroblast-like synoviocytes derived from osteoarthritis patients. The data suggest that cortisol-induced ZBTB16 acts as an intracellular modulator of glucocorticoid action by limiting GR-mediated activating effects and enhancing repressive effects. This mechanism may facilitate a return to the initial cellular state after (proinflammatory) stimulation and enhance GR's anti-inflammatory effects. This mechanism is similar to that of 'dissociated' GR ligands and may guide drug development that aims to reduce side effects while retaining the clinical benefits of glucocorticoid treatment.
Keywords: PLZF; cortisol; intracellular feedback; transactivation; transrepression.
Conflict of interest statement
OCM receives funding from Corcept Therapeutics, which develops GR modulators for clinical use. SG, EF, IK, LLK, JB, EvD, DE, RO, MS, and CJFB have nothing to disclose.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

