Dopamine and cortical neurons with different Parkinsonian mutations show variation in lysosomal and mitochondrial dysfunction
- PMID: 40541941
- PMCID: PMC12181412
- DOI: 10.1038/s41531-025-01048-2
Dopamine and cortical neurons with different Parkinsonian mutations show variation in lysosomal and mitochondrial dysfunction
Abstract
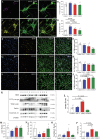
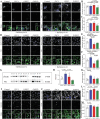
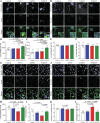
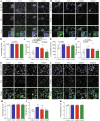
Mutations causing Parkinson's disease (PD) give diverse pathological phenotypes whose cellular correlates remain to be determined. Those with PRKN mutations have significantly earlier selective vulnerability of dopamine neurons, those with SNCA mutations have increased alpha-synuclein deposition, while those with LRRK2 mutations have additional deposition of tau. Yet all three mutation types are implicated in mitochondrial and/or lysosomal dysfunction. To compare cellular dysfunctions associated with these different pathological phenotypes, an unbiased high-content imaging platform was developed to assess both lysosomal and mitochondrial dysfunction, along with alpha-synuclein and tau protein deposition using induced pluripotent stem cell (iPSC) derived cortical and ventral midbrain neurons. Different PD mutations caused cell type specific dysfunctions, likely to impact on both selective neuronal vulnerability and the pathologies observed in PD. Comparison of dopamine neurons identified that both lysosomal and mitochondrial dysfunction were predominant with PRKN lof mutations, whereas SNCA A53T and LRRK2 R1441G mutations had increased tau deposition. In contrast, cortical neurons with SNCA and LRRK2 mutations both had mitochondrial and autophagy impairments without protein deposition, with LRRK2 cells additionally showing decreased glucocerebrosidase activity and increased alpha-synuclein phosphorylation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Kalia, L. V. & Lang, A. E. Parkinson’s disease. Lancet386, 896–912 (2015). - PubMed
-
- Halliday, G. et al. The progression of pathology in longitudinally followed patients with Parkinson’s disease. Acta Neuropathol.115, 409–415 (2008). - PubMed
-
- Schapira, A. H. V., Chaudhuri, K. R. & Jenner, P. Non-motor features of Parkinson disease. Nat. Rev. Neurosci.18, 435–450 (2017). - PubMed
-
- Lang, A. E. et al. Trial of Cinpanemab in Early Parkinson’s Disease. N. Engl. J. Med387, 408–420 (2022). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

