In-depth analysis of the mode of action of resveratrol: genome-wide characterization of G-quadruplex binding properties
- PMID: 40542407
- PMCID: PMC12180162
- DOI: 10.1186/s11658-025-00747-1
In-depth analysis of the mode of action of resveratrol: genome-wide characterization of G-quadruplex binding properties
Abstract
Background: Resveratrol (RSV) is one of the most studied and used biomolecules, for which many pharmacological effects targeting multiple tissues have been described. However, a common underlying mechanism driving its full pharmacological activity has not been described in detail. G-quadruplexes (G4s) are non-canonical nucleic acid structures found in regulatory genomic locations and involved in controlling gene transcription, telomere maintenance, or genome stability, among others. This study provides a genome-wide characterization of RSV G4-binding properties, explaining its multi-target traits.
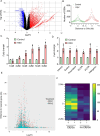
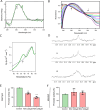
Methods: Immunofluorescence assays using a nucleolar and a G4-specific antibody were used to characterize RSV cellular effects on the nucleolus and G4 stabilization. DNA damage and cell cycle analyses were performed via western blot and flow cytometry. Breaks lLbeling In Situ and Sequencing (BLISS) was used to map double strand breaks (DSB) in response to treatment, and identify G4s targeted by RSV. mRNA sequencing was used to identify changes at the transcriptional level upon treatment and relate them to a direct targeting of G4s. Biophysical assays (circular dichroism, ultraviolet-visible [UV-Vis] titration, differential scanning calorimetry, and nuclear magnetic resonance) were used to characterize RSV-G4 interactions. Lastly, luciferase-based transcription assays were performed to confirm RSV-G4 interaction in vitro and its direct influence on gene expression.
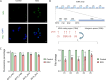
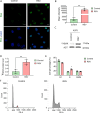
Results: In a cellular context, RSV treatment showed classic G4-ligand effects, such as nucleolar disassembly, inhibition of RNA polymerase I, DNA damage, and cell cycle arrest. RSV was shown to stabilize cellular G4s, which accumulated around double strand breaks in the promoters of differentially expressed genes. Upon treatment, G4 stabilization triggered DNA damage and controlled gene expression. The interaction between RSV and target G4s was confirmed in vitro by biophysical assays and through luciferase-based transcription assays.
Conclusions: A G4-dependent mode of action was demonstrated as the main mechanism underlying RSV pleiotropic effects, along with the identification of target genes and G4s. This in-depth analysis of the mode of action of RSV will be helpful to improve its therapeutic potential in a wide variety of health scenarios.
Keywords: DNA binding; G-quadruplex; Resveratrol; Secondary DNA structures; Small molecule.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: Authors declare that they have no competing interests.
Figures
Similar articles
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
References
-
- Takaoka M. Resveratrol, a new phenolic compound, from Veratrumgrandiflorum. J Chem Soc Jpn. 1939;60:1090–100.
-
- Tian B, Liu J. Resveratrol: a review of plant sources, synthesis, stability, modification and food application. J Sci Food Agric. 2020;100(4):1392–404. - PubMed
-
- Nunes S, Danesi F, Del Rio D, Silva P. Resveratrol and inflammatory bowel disease: the evidence so far. Nutr Res Rev. 2018;31(1):85–97. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

