LGALS3BP/90K suppresses porcine reproductive and respiratory syndrome virus replication by enhancing GP3 degradation and stimulating innate immunity
- PMID: 40542445
- PMCID: PMC12180180
- DOI: 10.1186/s13567-025-01556-2
LGALS3BP/90K suppresses porcine reproductive and respiratory syndrome virus replication by enhancing GP3 degradation and stimulating innate immunity
Abstract
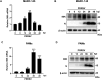
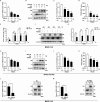
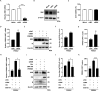
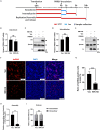
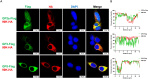
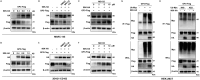
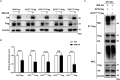
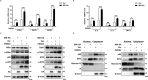
Porcine reproductive and respiratory syndrome virus (PRRSV) seriously affects the world pig industry, and existing vaccines are not enough to protect against PRRSV efficiently. Identifying host factors that inhibit PRRSV infection may provide potential targets for antiviral therapies. Galectin 3 binding protein (LGALS3BP), also referred to as 90K, is a multifunctional protein that plays significant roles in viral infections, but its role in PRRSV infection is not clearly defined. In this study, we found that 90K expression increased during PRRSV infection. Additionally, 90K inhibits PRRSV replication by restricting both viral RNA synthesis and viral assembly. Further studies showed that 90K can interact with various PRRSV structural proteins, such as GP2, GP3, and GP5. Moreover, 90K targets K217 in GP3 for K48-linked ubiquitination, facilitating GP3 degradation through the proteasomal pathway. Meanwhile, we found that 90K induces the expression of type I interferon (IFN) and inflammatory cytokines by activating NF-κB and IRF3. Overall, our findings suggest that 90K plays a negative role in PRRSV replication by inducing PRRSV-GP3 degradation and activating innate immune responses.
Keywords: LGALS3BP/90K; PRRSV; innate immunity; proteasomal degradation; viral replication.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare that they have no competing interests.
Figures

Similar articles
-
METTL3 regulates PRRSV replication by suppressing interferon beta through autophagy-mediated IKKε degradation.J Virol. 2025 Jul 22;99(7):e0009825. doi: 10.1128/jvi.00098-25. Epub 2025 Jun 23. J Virol. 2025. PMID: 40548751 Free PMC article.
-
The DEAD-box RNA helicase 27 negatively regulates the replication of porcine reproductive and respiratory syndrome virus by mediating GP2a autophagy degradation and inducing interferon-β production.Front Immunol. 2025 Jun 12;16:1587647. doi: 10.3389/fimmu.2025.1587647. eCollection 2025. Front Immunol. 2025. PMID: 40574840 Free PMC article.
-
Porcine reproductive and respiratory syndrome virus NSP5 exploited UBE2L6 to promote viral replication via antagonising host RLRs and ISGylation.Vet Res. 2025 Jul 3;56(1):136. doi: 10.1186/s13567-025-01558-0. Vet Res. 2025. PMID: 40611336 Free PMC article.
-
Coinfection with bacterial pathogens and genetic modification of PRRSV-2 for suppression of NF-κB and attenuation of proinflammatory responses.Virology. 2025 May;606:110484. doi: 10.1016/j.virol.2025.110484. Epub 2025 Mar 7. Virology. 2025. PMID: 40086205 Review.
-
Strategies and scheming: the war between PRRSV and host cells.Virol J. 2025 Jun 11;22(1):191. doi: 10.1186/s12985-025-02685-y. Virol J. 2025. PMID: 40500743 Free PMC article. Review.
References
-
- Artini M, Natoli C, Tinari N, Costanzo A, Marinelli R, Balsano C, Porcari P, Angelucci D, D’Egidio M, Levrero M, Iacobelli S (1996) Elevated serum levels of 90K/MAC-2 BP predict unresponsiveness to alpha-interferon therapy in chronic HCV hepatitis patients. J Hepatol 25:212–217 - PubMed
-
- Bosquillon DJL, Akbil B, Mhlekude B, Leyens J, Postmus D, Harnisch G, Jansen J, Schmidt ML, Aigner A, Pott F, Chua RL, Krist L, Gentile R, Muhlemann B, Jones TC, Niemeyer D, Fricke J, Keil T, Pischon T, Janke J, Conrad C, Iacobelli S, Drosten C, Corman VM, Ralser M, Eils R, Kurth F, Sander L, Goffinet C (2023) 90K/LGALS3BP expression is upregulated in COVID-19 but may not restrict SARS-CoV-2 infection. Clin Exp Med 23:3689–3700 - PMC - PubMed
-
- Creagh EM, O’Neill LA (2006) TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol 27:352–357 - PubMed
MeSH terms
Substances
Grants and funding
- ZR2024QC046/Natural Science Foundation of Shandong Province
- ZR2024QC167/Natural Science Foundation of Shandong Province
- ZR2021MC008/Natural Science Foundation of Shandong Province
- 31902259/National Natural Science Foundation of China
- SDAIT-08-07/Modern Agricultural Technology Industry System of Shandong province
LinkOut - more resources
Full Text Sources
Miscellaneous

