Origin and Evolution of Bacterial Periplasmic Force Transducers
- PMID: 40542537
- PMCID: PMC12204202
- DOI: 10.1093/molbev/msaf138
Origin and Evolution of Bacterial Periplasmic Force Transducers
Abstract
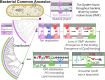
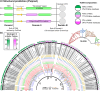
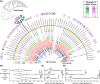
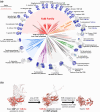
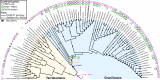
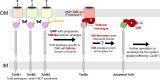
In double-membraned bacteria, non-equilibrium processes that occur at the outer membrane are typically coupled to the chemiosmotically energized inner membrane. TolA and TonB are homologous proteins which energetically couple inner membrane motor proteins to the essential processes of outer membrane stabilization and substrate import, respectively. The evolutionary trajectories of these proteins have been difficult to elucidate due to low-sequence conservation, yet they are thought to transduce force similarly. Here, this problem was addressed using structural prediction approaches to identify and annotate force transduction operons to trace their distribution and evolutionary origins. In the process, we identify a novel outer membrane-tethering system and a previously unknown family of monomeric force transducers. This approach revealed putative tolA genes, and thus the core organizational principles of the tol-pal operon throughout diverse bacterial taxa. We discovered that the α-helical structure of the periplasm-spanning domain II of TolA previously thought its hallmark, is anomalous amongst most Tol-Pal systems. This structure is mainly prevalent in γ-proteobacteria, likely in adaptation to their lifestyle. Comparison of Tol-Pal and Ton system distribution suggests that TolA emerged from a TonB paralogue and co-emerged with Pal, the outer membrane-tethering lipoprotein that functionalizes the Tol-Pal system. We also determined that TolB, the Pal-mobilizing protein, likely emerged from a family of outer membrane proteins; and CpoB, a periplasmic factor that coordinates peptidoglycan remodeling with cell division, was originally a lipoprotein present in the ancestral Tol-Pal system. The extensive conservation of the Tol-Pal system throughout Gracilicutes highlights its significance in bacterial cell biology.
Keywords: Tol-Pal system; Ton system; bacteria; bacterial cell envelope; force transduction; molecular motors.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

