PYF: a multi-functional algorithm for predicting production and optimizing metabolic engineering strategy in Escherichia coli microbial consortia
- PMID: 40542815
- PMCID: PMC12205937
- DOI: 10.1093/bib/bbaf295
PYF: a multi-functional algorithm for predicting production and optimizing metabolic engineering strategy in Escherichia coli microbial consortia
Abstract
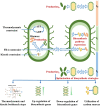
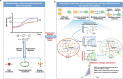
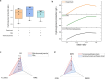
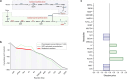
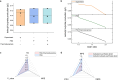
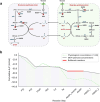
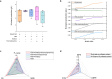
Simulating production in microbial consortia is crucial for optimizing metabolic engineering strategies to achieve high yields. However, existing algorithms for modeling polymicrobial metabolic fluxes, based on genome-scale metabolic networks, often overlook the conflicts and coordination between biosynthesis tasks and self-growth interests, leading to limited prediction accuracy. This study introduces the Polymicrobial cell factory Yield Forecasting (PYF) algorithm, which simulates the relationships between biosynthesis and growth more effectively by incorporating the expression degrees of biosynthesis pathways. PYF was shown to accurately predict the production of Escherichia coli-E. coli consortia under various scenarios, including mono-metabolite exchange, dual-carbon sources, and dual-metabolite exchange. The results revealed a mean relative error (MRE) of 0.106, an average determination coefficient of 0.883, and an average hypothesis testing parameter of 0.930 between predicted and experimental productions. Compared with the recent metabolic simulation algorithm, PYF reduced the MRE by ~61.6%. PYF is adaptable and enables accurate simulation even without enzyme catalytic data. Meanwhile, PYF rapidly analyzed and optimized metabolic engineering strategies through sensitivity analysis. By eliminating the need for specialized division and integration of polymicrobial metabolic networks, PYF greatly simplifies the simulation process, offering a novel approach for predicting and enhancing production in microbial consortia.
Keywords: biosynthesis pathway expression degree; metabolic engineering strategy optimization; metabolic simulation algorithm; microbial consortium; production prediction.
© The Author(s) 2025. Published by Oxford University Press.
Figures

Similar articles
-
Metabolic engineering of Escherichia coli for high-yield dopamine production via optimized fermentation strategies.Appl Environ Microbiol. 2025 Jun 18;91(6):e0015925. doi: 10.1128/aem.00159-25. Epub 2025 May 8. Appl Environ Microbiol. 2025. PMID: 40338089 Free PMC article.
-
Integration of metatranscriptomics data improves the predictive capacity of microbial community metabolic models.ISME J. 2025 Jan 2;19(1):wraf109. doi: 10.1093/ismejo/wraf109. ISME J. 2025. PMID: 40448581 Free PMC article.
-
Integrating Gut Microbiome and Metabolomics with Magnetic Resonance Enterography to Advance Bowel Damage Prediction in Crohn's Disease.J Inflamm Res. 2025 Jun 11;18:7631-7649. doi: 10.2147/JIR.S524671. eCollection 2025. J Inflamm Res. 2025. PMID: 40535353 Free PMC article.
-
Rational construction of synthetic consortia: Key considerations and model-based methods for guiding the development of a novel biosynthesis platform.Biotechnol Adv. 2024 May-Jun;72:108348. doi: 10.1016/j.biotechadv.2024.108348. Epub 2024 Mar 24. Biotechnol Adv. 2024. PMID: 38531490 Review.
-
A Comprehensive Review of l-Theanine Production in Escherichia coli: The Recent Progress, Metabolic Engineering Strategies, and Future Prospects.ACS Synth Biol. 2025 Jul 18;14(7):2433-2444. doi: 10.1021/acssynbio.5c00291. Epub 2025 Jul 4. ACS Synth Biol. 2025. PMID: 40613122 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

