ATF4 in proximal tubules modulates kidney function and modifies the metabolome
- PMID: 40542844
- PMCID: PMC12343659
- DOI: 10.1007/s00109-025-02559-4
ATF4 in proximal tubules modulates kidney function and modifies the metabolome
Abstract
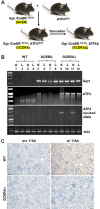
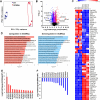
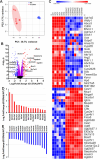
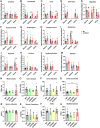
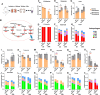
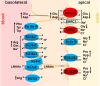
Activating transcription factor 4 (ATF4) is a transcription factor that mediates the response to stress at the cellular, tissue, and organism level. We deleted the gene encoding ATF4 in the proximal tubules of the mouse kidney by using a temporal and cell type-specific approach. We show that ATF4 plays a major role in regulating the transcriptome and proteome, which, in turn, influences the metabolome and kidney functions. Genome-wide transcriptomics and single-plot, solid-phase-enhanced sample preparation (SP3)-proteomics studies reveal that ATF4 deletion changes more than 30% of transcripts and, similarly, corresponding proteins in the proximal tubules. Gene Set Enrichment Analysis indicates major changes in transporters, including amino acid transporters. Metabolomic analyses show that these changes in transporters are associated with altered profiles of amino acids in the blood, kidney, and urine. Stable isotope glutamine tracing in primary tubule cells isolated from kidney cortices confirms that ATF4 regulates glutamine transport and metabolism. We suggest that even in the absence of additional stresses, such as kidney injury, ATF4 in the proximal tubules modulates both retention of specific nutrients and excretion of catabolic products like creatinine to maintain normal kidney function. KEY MESSAGES: Activating transcription factor 4 (ATF4) deletion changed more than 30% of genome-wide transcripts and corresponding proteins in the proximal tubules. One set of the profound changes occurred in amino acid transporters and Slc22 family transporters. Changes in transporters were accompanied by altered profiles of amino acids and wastes in the blood, kidney, and urine. ATF4 in the kidney proximal tubules plays a key role in regulating both the reabsorption of nutrients and the excretion of wastes.
Keywords: Activating transcription factor 4 (ATF4); Kidney proximal tubules; Metabolomics; Proteomics; Transcriptomics; Transporters.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Declarations. Ethics approval: All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of WCMC. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Deletion of the transcription factor ATF4 in a model of clear cell renal cell carcinoma.Neoplasia. 2025 Aug;66:101188. doi: 10.1016/j.neo.2025.101188. Epub 2025 Jun 4. Neoplasia. 2025. PMID: 40472739 Free PMC article.
-
ATF4 promotes glutaminolysis and glycolysis in colorectal cancer by transcriptionally inducing SLC1A5.Acta Biochim Biophys Sin (Shanghai). 2024 Dec 17;57(7):1093-1105. doi: 10.3724/abbs.2024226. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39696988
-
Kir5.1 regulates Kir4.2 expression and is a key component of the 50-pS inwardly rectifying potassium channel in basolateral membrane of mouse proximal tubules.Am J Physiol Renal Physiol. 2025 Feb 1;328(2):F248-F257. doi: 10.1152/ajprenal.00178.2024. Epub 2025 Jan 2. Am J Physiol Renal Physiol. 2025. PMID: 39745541 Free PMC article.
-
Energy-Dependent Urea Transports in Mammals and their Functional Consequences.Subcell Biochem. 2025;118:193-228. doi: 10.1007/978-981-96-6898-4_10. Subcell Biochem. 2025. PMID: 40637983 Review.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
-
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10:307–318. 10.1038/nrm2672 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

