Swine Xenografts Share Few Predicted Indirectly Recognisable SLA-Derived Epitopes With HLA-Derived Epitopes From Human Kidney Grafts
- PMID: 40543909
- PMCID: PMC12182435
- DOI: 10.1111/tan.70291
Swine Xenografts Share Few Predicted Indirectly Recognisable SLA-Derived Epitopes With HLA-Derived Epitopes From Human Kidney Grafts
Abstract
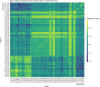
Swine-derived kidneys are a promising alternative organ source for transplantation, but compatibility in the major histocompatibility complex remains an immunological barrier. Furthermore, in repeat transplantations, CD4+ memory T cells can lead to a more rapid immune response against repeated exposure to the same antigens. Several studies have shown that HLA and SLA proteins share overlapping B cell epitopes due to structural or electrostatic similarities, but the role of overlapping T cell epitopes has not been fully explored. This study aims to computationally analyse the potential risk of memory T cell activation in subsequent human-after-swine and swine-after-human transplantation by evaluating shared T cell epitopes between the two graft sources. We show that while HLA and SLA demonstrate striking structural similarities, their linear protein sequences are very distinct, which translates to disparate HLA- and SLA-derived peptidomes and T cell epitopes. By applying the PIRCHE-II Tmem analysis to a simulated panel of recipients receiving repeat transplantations from a human kidney and from a swine xenograft, we observed a median of 1 shared T cell epitope in the cross-species context, compared to a median of 17 shared between two human-derived kidneys. This suggests that a swine xenograft exposes a low risk of T cell memory against a later human donor, and that xenotransplantation may provide an opportunity to receive a graft for highly HLA-sensitised recipients.
Keywords: PIRCHE; SLA; memory; swine; xenotransplantation.
HLA: Immune Response Genetics© 2025 The Author(s). HLA: Immune Response Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The UMC Utrecht has filed patent applications on the prediction of an alloimmune response against mismatched HLA. E.S. is listed as an inventor on these patents. B.M.M. and M.N. are employed by PIRCHE AG, which publishes the PIRCHE web portal.
Figures

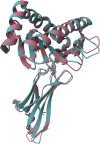


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

