Possible Acquisition and Molecular Evolution of vpu Genes Inferred from Comprehensive Sequence Analysis of Human and Simian Immunodeficiency Viruses
- PMID: 40544231
- PMCID: PMC12354576
- DOI: 10.1007/s00239-025-10256-6
Possible Acquisition and Molecular Evolution of vpu Genes Inferred from Comprehensive Sequence Analysis of Human and Simian Immunodeficiency Viruses
Abstract
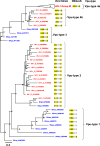
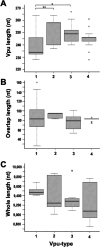
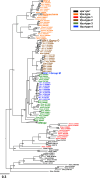
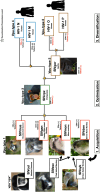
Vpu, an accessory protein of human immunodeficiency virus-1 (HIV-1), plays a crucial role in viral particle production and significantly contributes to HIV virulence. However, the evolution of the vpu gene remains poorly understood. We conducted a computational analysis of approximately 39,000 simian immunodeficiency virus (SIV) and HIV sequences, focusing on 141 representative Vpu proteins. Phylogenetic analysis classified the SIV and HIV strains into four major types based on their Vpu proteins: Vpu-type 1 (ancestral, found in SIVs such as SIVmon and SIVgsn), Vpu-type 2 (SIVgor and HIV-1 group O), Vpu-type 3 (SIVcpz), and Vpu-type 4 (HIV-1 group M and N). Notably, Vpu-type 1 exhibited variability in gene length, genome length, and the overlap between vpu and env compared with other Vpu-types. A phylogenetic tree was constructed using 426 nucleotide sequences from HIV-1, HIV-2, and SIVs focusing on the region between the pol and env genes. Vpu-type 1 was closely clustered with SIVasc and SIVsyk, lacking both vpu and vpx. The similarities observed between vpu and genes such as vpr and env suggest that vpu originated within the SIV genome. In addition, a phylogenetic tree constructed from 252 Vpu-type 4a sequences from the HIV pandemic strain and 135 sequences of circulating recombinant forms of HIV-1 revealed 18 distinct protein subtypes, exceeding the number of previously recognized subtypes. The systematic analysis of the sequences from large datasets has enabled a detailed characterization of the transition states of vpu, enhancing our understanding of the processes driving viral diversity.
Keywords: Bioinformatics; HIV-1; Molecular evolution; Phylogenetic tree; SIV; Vpu.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of Interest: The authors declare that they have no conflicts of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

