Uncovering encrypted antimicrobial peptides in health-associated Lactobacillaceae by large-scale genomics and machine learning
- PMID: 40544303
- PMCID: PMC12181893
- DOI: 10.1186/s40168-025-02145-3
Uncovering encrypted antimicrobial peptides in health-associated Lactobacillaceae by large-scale genomics and machine learning
Abstract
Background: Antimicrobial peptides (AMPs) are well known for their broad-spectrum activity and have shown great promise in addressing the antibiotic-resistant crisis. The Lactobacillaceae family, recognized for its health-promoting effects in humans, represents a valuable source of novel AMPs. However, the global prevalence and distribution of AMPs within Lactobacillaceae remains largely unknown, which limits the efficient discovery and development of novel AMPs.
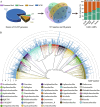
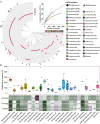
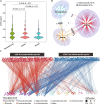
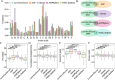
Results: We analyzed all available genomes (10,327 genomes), encompassing 38 genera and 515 species, to investigate the biosynthetic potential (indicated by the number of AMP sequences in the genome) of AMP in the Lactobacillaceae family. We demonstrated Lactobacillaceae species had ubiquitous (69.90%) biosynthetic potential of AMPs. Overall, 9601 AMPs were identified, clustering into 2092 gene cluster families (GCFs), which showed strong interspecies specificity (95.27%), intraspecies heterogeneity (93.31%), and habitat uniqueness (95.83%), that greatly expanded on the AMP sequence landscape. Novelty assessment indicated that 1516 GCFs (72.47%) had no similarity to any known AMPs in existing databases. Machine learning predictions suggested that novel AMPs from Lactobacillaceae possessed strong antimicrobial potential, with 664 GCFs having an additive minimum inhibitory concentration (MIC) below 100 μM. We randomly synthesized 16 AMPs (with predicted MIC < 100 μM) and identified 10 AMPs exhibiting varied-spectrum activity against 11 common pathogens. Finally, we identified one Lactobacillus delbrueckii-originated AMP (delbruin_1) having broad-spectrum (all 11 pathogens) and high antimicrobial activity (average MIC = 38.56 µM), which proved its potential as a clinically viable antimicrobial agent.
Conclusions: We uncovered the global prevalence of AMPs in Lactobacillaceae and proved that Lactobacillaceae is an untapped and invaluable source of novel AMPs to combat the antibiotic-resistance crisis. Meanwhile, we provided a machine learning-guided framework for AMP discovery, offering a scalable roadmap for identifying novel AMPs not only in Lactobacillaceae but also in other organisms. Video Abstract.
Keywords: Lactobacillaceae; Antibiotic resistance; Antimicrobial peptides; Genome mining; Machine learning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
- GZC20240728/Postdoctoral Fellowship Program of CPSF
- 2024ZB271/The Jiangsu Funding Program for Excellent Postdoctoral Talent
- xjnkywdzc-2024001-10/Project of Fund for Stable Support to Agricultural Sci-Tech Renovation
- No. 111-2-06/Priority Academic Program Development of Jiangsu Higher Education Institutions
- 32172175/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

