Uncovering injury-specific proteomic signatures and neurodegenerative risks in single and repetitive traumatic brain injury
- PMID: 40545497
- PMCID: PMC12183312
- DOI: 10.1038/s41392-025-02286-9
Uncovering injury-specific proteomic signatures and neurodegenerative risks in single and repetitive traumatic brain injury
Abstract
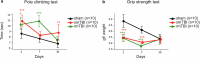
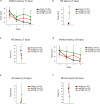
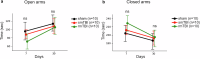
Traumatic brain injury (TBI) is a major public health concern associated with an increased risk of neurodegenerative diseases including Alzheimer's disease (AD), Parkinson's disease (PD), and chronic traumatic encephalopathy, yet the underlying molecular mechanisms in repetitive TBI remain poorly defined. This study investigates proteomic and behavioral changes following single and repetitive mild TBI in a mouse model, focusing on molecular alterations in the cortex and hippocampus across acute (48 h) and subacute (1 week) stages. Using shotgun proteomics and bioinformatics approaches, including weighted gene co-expression network analysis (WGCNA) and machine learning, we analyzed the proteomic landscapes of TBI-affected brain regions including the hippocampus and the cortex. We assessed motor and cognitive outcomes at 2-, 7-, and 30-days post-injury to explore functional impairments associated with observed molecular changes. Our findings reveal spatio-temporal injury- and time-specific proteomic changes, with a single TBI promoting neuroprotective and repair mechanisms, while repetitive TBI exacerbating neuronal damage and synaptic deficits in the hippocampus. Key deregulated proteins, including Apoa1, ApoE, Cox6a1, and Snca, were linked to neurodegenerative pathways, suggesting molecular connections between TBI and diseases like AD and PD. Behavioral assessments indicated that repetitive TBI significantly impaired motor and cognitive functions, with recovery in motor function by day 30, whereas cognitive deficits persisted. This study provides a detailed analysis of the proteomic and behavioral consequences of TBI, identifying molecular networks as potential biomarkers or therapeutic targets for mitigating long-term cognitive decline associated with repetitive head trauma. These findings underscore the importance of mitochondrial and synaptic integrity in TBI response and suggest that targeting these pathways could reduce neurodegenerative risk following repetitive TBI.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

