Analysis of IDH1 and IDH2 mutations as causes of the hypermethylator phenotype in colorectal cancer
- PMID: 40545636
- PMCID: PMC12337813
- DOI: 10.1002/path.6446
Analysis of IDH1 and IDH2 mutations as causes of the hypermethylator phenotype in colorectal cancer
Abstract
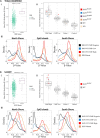
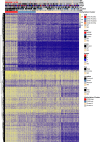
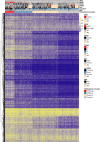
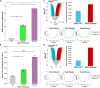
The CpG island methylator phenotype (CIMP) occurs in many colorectal cancers (CRCs). CIMP is closely associated with global hypermethylation and tends to occur in proximal tumours with microsatellite instability (MSI), but its origins have been obscure. A few CRCs carry oncogenic (gain-of-function) mutations in isocitrate dehydrogenase IDH1. Whilst IDH1 is an established CRC driver gene, the low frequency of IDH1-mutant CRCs (about 0.5%) has meant that the effects and molecular covariates of those mutations have not been established. We first showed computationally that IDH2 is also a CRC driver. Using multiple public and in-house CRC datasets, we then identified IDH mutations at the hotspots (IDH1 codons 132 and IDH2 codons 140 and 172) frequently mutated in other tumour types. Somatic IDH mutations were associated with BRAF mutations and expression of mucinous/goblet cell markers, but not with KRAS mutations or MSI. All IDH-mutant CRCs were CIMP-positive, mostly at a high level. Cell and mouse models showed that IDH mutation was plausibly causal for DNA hypermethylation. Whilst the aetiology of hypermethylation generally remains unexplained, IDH-mutant tumours did not form a discrete methylation subcluster, suggesting that different underlying mechanisms can converge on similar final methylation phenotypes. Although further analysis is required, IDH mutations may be the first cause of hypermethylation to be identified in a common cancer type, providing evidence that CIMP and DNA methylation represent more than aging-related epiphenomena. Cautious exploration of mutant IDH inhibitors and DNA demethylating agents is suggested in managing IDH-mutant CRCs. © 2025 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: DNA methylation; cancer driver genes; cancer genomics; colorectal cancer; epigenetics; isocitrate dehydrogenase.
© 2025 The Author(s). The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

