Engineered Marine Biofilms for Ocean Environment Monitoring
- PMID: 40545707
- PMCID: PMC12281610
- DOI: 10.1021/acssynbio.5c00192
Engineered Marine Biofilms for Ocean Environment Monitoring
Abstract
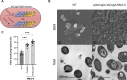
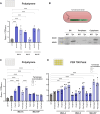
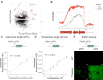
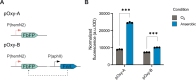
Marine bacteria offer a promising alternative for developing Engineered Living Materials (ELMs) tailored to marine applications. We engineered Dinoroseobacter shibae to increase its surface-associated growth and develop biosensors for ocean environment monitoring. By fusing the endogenous extracellular matrix amyloidogenic protein CsgA with mussel foot proteins, we significantly increased D. shibae biofilm formation. Additionally, D. shibae was engineered to express the tyrosinase enzyme to further enhance microbial attachment through post-translational modifications of tyrosine residues. By exploiting D. shibae's natural genetic resources, two environmental biosensors were created to detect temperature and oxygen. These biosensors were coupled with a CRISPR-based recording system to store transient gene expression in stable DNA arrays, enabling long-term environmental monitoring. These engineered strains highlight D. shibae's potential in advancing marine microbiome engineering for innovative biofilm applications, including the development of natural, self-renewing biological adhesives, environmental sensors, and "sentinel" cells equipped with CRISPR-recording technology to capture and store environmental signals.
Keywords: Dinoroseobacter shibae; ELMs; biofilm; biosensors; marine bacteria; surface colonization.
Figures
Similar articles
-
Microbial communities associated with plastic fishing nets: diversity, potentially pathogenic and hydrocarbon degrading bacteria.Sci Rep. 2025 Jul 2;15(1):22877. doi: 10.1038/s41598-025-06033-6. Sci Rep. 2025. PMID: 40594211 Free PMC article.
-
Evolutionary history and association with seaweeds shape the genomes and metabolisms of marine bacteria.mSphere. 2025 Jun 25;10(6):e0099624. doi: 10.1128/msphere.00996-24. Epub 2025 Jun 2. mSphere. 2025. PMID: 40454874 Free PMC article.
-
Liposome-encapsulated antibiotics and biosurfactants: an effective strategy to boost biofilm eradication in cooling towers.Microb Cell Fact. 2025 Jun 18;24(1):135. doi: 10.1186/s12934-025-02746-5. Microb Cell Fact. 2025. PMID: 40533735 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Hybrid closed-loop systems for managing blood glucose levels in type 1 diabetes: a systematic review and economic modelling.Health Technol Assess. 2024 Dec;28(80):1-190. doi: 10.3310/JYPL3536. Health Technol Assess. 2024. PMID: 39673446 Free PMC article.
References
-
- Zhang C., Huang J., Zhang J., Liu S., Cui M., An B., Wang X., Pu J., Zhao T., Fan C., Lu T. K., Zhong C.. Engineered Bacillus Subtilis Biofilms as Living Glues. Mater. Today. 2019;28:40–48. doi: 10.1016/j.mattod.2018.12.039. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

