Mitochondrial proteome landscape unveils key insights into melanoma severity and treatment strategies
- PMID: 40545870
- PMCID: PMC12183497
- DOI: 10.1002/cncr.35897
Mitochondrial proteome landscape unveils key insights into melanoma severity and treatment strategies
Abstract
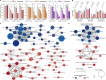
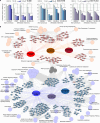
Background: Melanoma, the deadliest form of skin cancer, exhibits resistance to conventional therapies, particularly in advanced and metastatic stages. Mitochondrial pathways, including oxidative phosphorylation and mitochondrial translation, have emerged as critical drivers of melanoma progression and therapy resistance. This study investigates the mitochondrial proteome in melanoma to uncover novel therapeutic vulnerabilities.
Methods: Quantitative proteomics was performed on 151 melanoma-related samples from a prospective cohort and postmortem tissues. Differential expression analysis identified mitochondrial proteins linked to disease aggression and treatment resistance. Functional enrichment analyses and in vitro validation using mitochondrial inhibitors were conducted to evaluate therapeutic potential.
Results: Mitochondrial translation and oxidative phosphorylation (OXPHOS) were significantly upregulated in aggressive melanomas, particularly in BRAF-mutant and metastatic tumors. Inhibition of mitochondrial pathways using antibiotics (doxycycline, tigecycline, and azithromycin) and OXPHOS inhibitors (VLX600, IACS-010759, and BAY 87-2243) demonstrated dose-dependent antiproliferative effects in melanoma cell lines, sparing noncancerous melanocytes. These treatments disrupted mitochondrial function, suppressed key metabolic pathways, and induced apoptosis, highlighting the clinical relevance of targeting these pathways.
Conclusions: This study reveals mitochondrial pathways as critical drivers of melanoma progression and resistance, providing a rationale for targeting mitochondrial translation and OXPHOS in advanced melanoma. Combining mitochondrial inhibitors with existing therapies could overcome treatment resistance and improve patient outcomes.
Keywords: BRAF mutation; MCM complex; melanoma; mitochondrial metabolism; mitochondrial proteome; mitoribosomes; oxidative phosphorylation; proteomics.
© 2025 The Author(s). Cancer published by Wiley Periodicals LLC on behalf of American Cancer Society.
Conflict of interest statement
Fábio C. S. Nogueira reports grant and/or contract funding from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro. Jéssica Guedes reports fees for professional activities from Lund University. The other authors declare no conflicts of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

