Extracellular Vesicle Proteome Analysis Improves Diagnosis of Recurrence in Triple-Negative Breast Cancer
- PMID: 40545963
- PMCID: PMC12183389
- DOI: 10.1002/jev2.70089
Extracellular Vesicle Proteome Analysis Improves Diagnosis of Recurrence in Triple-Negative Breast Cancer
Abstract
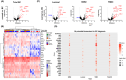
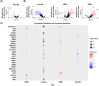
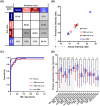
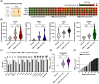
We explored the diagnostic utility of tumor-derived extracellular vesicles (tdEVs) in breast cancer (BC) by performing comprehensive proteomic profiling on plasma samples from 130 BC patients and 40 healthy controls (HC). Leveraging a microfluidic chip-based isolation technique optimized for low plasma volume and effective contaminant depletion, we achieved efficient enrichment of tdEVs. Proteomic analysis identified 26 candidate biomarkers differentially expressed between BC patients and HCs. To enhance biomarker selection robustness, we implemented a hybrid machine learning framework integrating LsBoost, convolutional neural networks, and support vector machines. Among the identified candidates, four EV proteins. ECM1, MBL2, BTD, and RAB5C. not only exhibited strong discriminatory performance, particularly for triple-negative breast cancer (TNBC), but also demonstrated potential relevance to disease recurrence, providing prognostic insights beyond initial diagnosis. Receiver operating characteristic (ROC) curve analysis demonstrated high diagnostic accuracy with an area under the curve (AUC) of 0.924 for BC and 0.973 for TNBC, as determined by mass spectrometry. These findings were further substantiated by immuno assay validation, which yielded an AUC of 0.986 for TNBC. Collectively, our results highlight the potential of EV proteomics as a minimally invasive, blood-based platform for both accurate detection and recurrence risk stratification in breast cancer and its aggressive subtypes, offering promising implications for future clinical applications.
Keywords: diagnosis; machine learning; microfluidics; proteomic analysis; triple‐negative breast cancer; tumour derived extracellular vesicles.
© 2025 The Author(s). Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Bandu, R. , Oh J. W., and Kim K. P.. 2024. “Extracellular Vesicle Proteins as Breast Cancer Biomarkers: Mass Spectrometry‐based Analysis.” Proteomics 24: 2300062. - PubMed
-
- Baptistella, A. R. , Landemberger M. C., Dias M. V., et al. 2019. “Rab5C Enhances Resistance to Ionizing Radiation in Rectal Cancer.” Journal of Molecular Medicine (Berlin) 97: 855–869. - PubMed
MeSH terms
Substances
Grants and funding
- C-2023-0006/Severance Hospital Research Fund for Clinical Excellence grant
- 2020003030007/Korea Environmental Industry and Technology Institute
- 1485019157/Korea Environmental Industry and Technology Institute
- 2014R1A6A9064166/National Research Foundation of Korea
- 2021R1A2C3011254/National Research Foundation of Korea
LinkOut - more resources
Full Text Sources
Miscellaneous

